Sanger Sequencing to Illumina Sequencing: A Comparative Overview
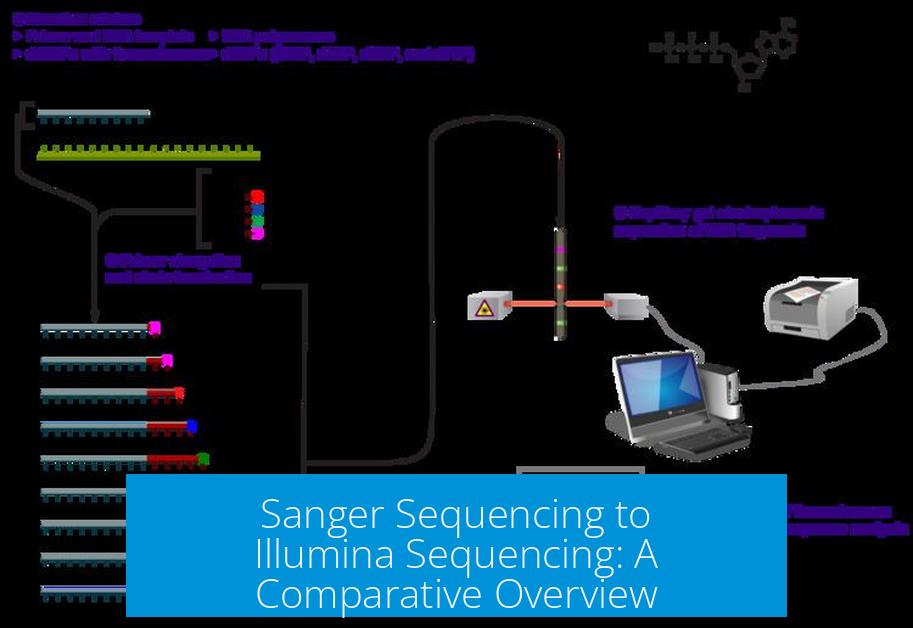
Sanger sequencing and Illumina sequencing represent two distinct methodologies in DNA sequencing, differing significantly in sample requirements, library preparation, throughput, and application scope. Transitioning from Sanger to Illumina sequencing involves adapting to higher quality DNA inputs, more complex library preparation with indexing, and massively parallel sequencing that enables multiplexing and higher data output.
1. Differences in Sample Isolation and DNA Quality Requirements
Both sequencing techniques begin with DNA isolation and purification. For Illumina sequencing, the quality of the sample is paramount. While the same extraction methods applicable to Sanger sequencing can be used, Illumina sequencing demands higher quality, intact DNA that is free from impurities and degradation.
- DNA Quality: Illumina sequencing (Next-Generation Sequencing, NGS) requires high-quality DNA. Contaminants or degradation reduce efficiency and data accuracy.
- DNA Quantification: Accurate quantification is critical. Fluorometric methods (e.g., Qubit or PicoGreen) provide reliable DNA concentration measurements by selectively quantifying double-stranded DNA.
- Purification Post-Extraction: Certain extraction protocols such as Quick Extract necessitate further purification. A common practice is Single-stranded Paramagnetic bead (SPRI) cleanup, eluting DNA in low EDTA buffer or water to avoid interference in downstream steps.
In contrast, Sanger sequencing tolerates slightly lower quality DNA because it sequences one fragment at a time and requires less complex library preparation.
2. Library Preparation: From Simple to Sophisticated
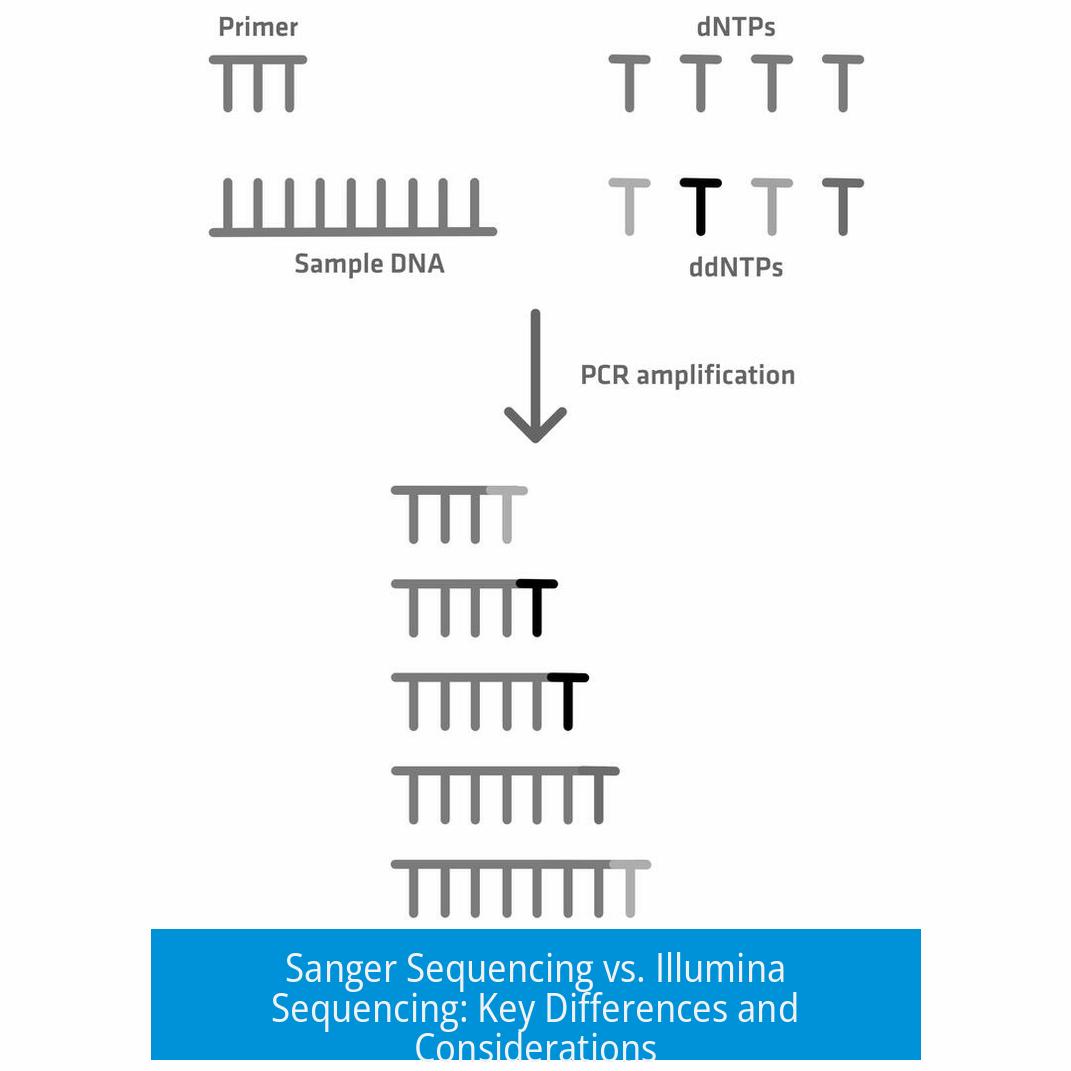
The most notable methodological difference lies in library preparation.
- Sanger Sequencing Library Prep: Typically involves amplification of a single DNA fragment with primers and direct sequencing of the amplified product. There is no need for adapters or indexing.
- Illumina Library Prep: Requires fragmenting the DNA, repairing ends, and ligating specialized adapters containing flow cell binding sites and indexes. Amplification with primers carrying Illumina-specific adapters is essential.
Illumina’s workflow supports multiplexing through indexing primers, allowing multiple samples to be pooled into a single sequencing run. This pooling capacity is a major advantage over Sanger sequencing, which sequences individual samples separately.
3. Sequencing Technology and Throughput Differences
Sanger sequencing uses chain termination chemistry and capillary electrophoresis to read DNA sequences. It produces accurate, long reads (up to ~900 base pairs) but has low throughput and high cost per base.
Illumina sequencing employs sequencing-by-synthesis (SBS) technology. It generates billions of short reads (typically 75-300 base pairs) simultaneously. This massive parallelism reduces cost, increases speed, and allows for applications such as whole-genome sequencing, exome sequencing, and RNA-seq.
Because Illumina short reads are comparatively shorter than Sanger reads, assembly complexity may increase for certain applications, but increased coverage depth compensates for read length.
4. Intermediate Technologies: Oxford Nanopore Sequencing
Between Sanger and Illumina sequencing lies Oxford Nanopore technology. It offers real-time, long-read sequencing but with different error profiles. Nanopore sequencing provides an intermediate step towards high-throughput sequencing with longer reads, useful for applications requiring long-range genomic information not well resolved by short reads.
5. Considerations for Long-Read Sequencing
When considering long-read sequencing, a high molecular weight (HMW) DNA extraction protocol is essential to preserve long fragments. Illumina’s “long-read” capabilities are limited and not widely accessible.
For long-read needs, Oxford Nanopore or Pacific Biosciences (PacBio) are preferable. Illumina remains dominant in short-read, high-accuracy applications.
6. Recommendations for Services and Workflow Setup
Many researchers use commercial services such as Azenta/Genewiz for NGS runs. These providers offer expertise in library preparation, sequencing, and data analysis, simplifying workflow complexity.
When planning to transition from Sanger to Illumina sequencing, considering sample quality, library prep requirements, indexing for multiplexing, and data analysis capabilities is critical.
Summary of Key Differences and Considerations
| Aspect | Sanger Sequencing | Illumina Sequencing |
|---|---|---|
| DNA Input Quality | Moderate tolerance | High-quality, purified, non-degraded DNA required |
| DNA Quantification Method | Sufficient with spectrophotometric methods | Fluorometric (Qubit/PicoGreen) is necessary |
| Library Preparation | Simple PCR amplification, no adapters needed | Complex protocol with fragmentation, adapter ligation, indexing primers |
| Sequencing Read Length | Long (around 900 bp) | Short (75–300 bp typical) |
| Throughput | Low, single sample per run | High, multiplexing dozens to hundreds of samples |
| Applications | Targeted sequencing, single gene analysis | Whole genomes, transcriptomes, complex genetic analysis |
| Cost | Higher per base | Lower per base with scale |
Practical Tips for Transition
- Use the same DNA isolation methods when possible but confirm DNA integrity and purity before Illumina library prep.
- Measurement of DNA quantity should rely on fluorometric assays to avoid overestimation from RNA or contaminants.
- Perform SPRI cleanup after extraction if necessary to remove inhibitors.
- Prepare libraries using Illumina-specific adapters and indexing primers to enable multiplexing capabilities.
- Engage professional sequencing services for best results if unfamiliar with NGS protocols.
Conclusion
Sanger and Illumina sequencing serve complementary roles in molecular biology. Sanger excels at precise, single-sample sequencing with long reads. Illumina provides high-throughput, short-read data well suited for genome-wide applications. Transitioning successfully involves ensuring superior DNA quality, adopting complex library prep, and leveraging indexing to maximize sequencing efficiency.
Key Takeaways
- Illumina sequencing requires higher quality DNA than Sanger sequencing.
- Fluorometric DNA quantification is essential for Illumina workflows.
- Illumina library preparation introduces adapters and indexing primers for multiplexing.
- Sanger sequencing reads are longer but throughput is low compared to Illumina.
- Oxford Nanopore sequencing acts as an intermediate sequencing technology.
- Professional NGS services can streamline the transition from Sanger to Illumina sequencing.
What are the main differences in sample preparation between Sanger and Illumina sequencing?
Both use similar isolation and purification methods. However, Illumina sequencing demands higher quality, non-degraded DNA as input. Post-extraction cleanup like SPRI is recommended for Illumina to ensure purity.
Why is DNA quantification important when transitioning from Sanger to Illumina sequencing?
Illumina sequencing requires precise DNA quantification using fluorometric methods such as Qubit or PicoGreen. Spectrophotometric methods like NanoDrop can overestimate DNA due to RNA contamination, affecting library prep and sequencing results.
How does library preparation differ between Sanger and Illumina sequencing?
Illumina library prep involves attaching specific adapters and amplification primers. It also supports multiplexing through indexing, allowing many samples to be pooled in a single run. Sanger sequencing lacks these complex library requirements.
Is Oxford Nanopore sequencing a suitable intermediate step between Sanger and Illumina?
Yes, Oxford Nanopore can act as a bridge technology. It offers longer reads than Illumina but differs in error profiles and workflows. Its role is often to complement or supplement next-generation sequencing methods.
Why is long-read DNA extraction important and is Illumina suitable for long reads?
Long-read sequencing requires high molecular weight DNA extraction for quality results. Currently, Illumina’s long-read technology is not widely available, so alternative platforms are preferred for long-read applications.




Leave a Comment