16S rRNA PCR Help: Key Considerations and Troubleshooting
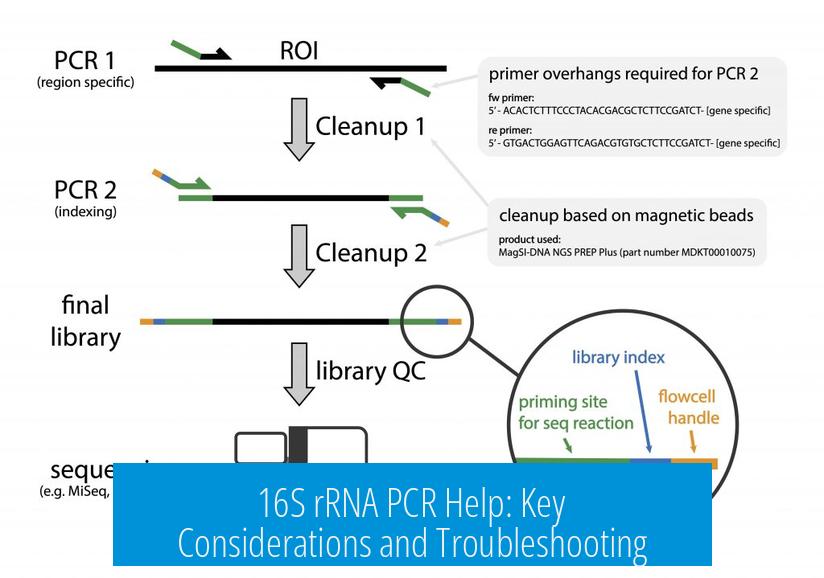
Effective 16S rRNA PCR depends on precise primer design, correct target gene selection, sample quality, and validating amplification through controls. Issues in any of these areas can cause amplification failure or misleading results.
1. Primer Design Challenges for 16S rRNA PCR
Designing primers for 16S rRNA requires careful attention to polymorphisms in the target sequences. Single nucleotide polymorphisms (SNPs) within 16S regions can disrupt primer binding, especially if they occur near the primer’s 3′ end. Mismatches at this terminal region often prevent primer extension and result in PCR failure.
To improve amplification success, primers with degenerate bases at the 3′ end can accommodate sequence variation. Identifying polymorphic sites in the organism of interest and incorporating degeneracy at these positions strengthens primer universality.
2. Choosing the Appropriate Ribosomal Target Gene
16S rRNA gene targets prokaryotic organisms, including bacteria and archaea. For eukaryotic species like Vetigastropoda mollusks, 18S rRNA primers are more suitable. Therefore, selecting primers consistent with the biology of the organism is crucial.
Additionally, cross-amplification with internal transcribed spacer (ITS) regions, such as ITS-2, may occur in eukaryotic PCR assays. This crosstalk can cause non-specific products, so one must carefully design and validate primers when targeting eukaryotic ribosomal genes.
3. Sample Quality and DNA Quantification
DNA concentration measured by NanoDrop serves as a guideline rather than an absolute threshold. Samples showing less than 10 ng/μL DNA may still yield successful PCR and sequencing results. Quality and integrity of DNA, along with appropriate dilution, often play a more significant role than raw concentration values alone.
4. Basic PCR Troubleshooting
Before testing samples, always validate primers using positive control templates known to amplify the target region. This step confirms primer functionality and identifies problems unrelated to sample DNA quality, such as reagent issues or suboptimal cycling conditions.
Summary of Key Points
- SNPs near primer 3′ ends can disrupt 16S PCR; introduce degeneracy to improve universality.
- Use 16S primers for prokaryotes; switch to 18S primers for eukaryotes to prevent misamplification.
- Low DNA concentration by NanoDrop does not necessarily prevent successful PCR.
- Validate primers with positive controls to confirm they can amplify target sequences.
What impact do SNPs have on the effectiveness of universal 16S rRNA primers?
Single nucleotide polymorphisms (SNPs) in the 16S region can cause mismatches with universal primers. Especially mismatches at the primer’s 3′ end reduce binding and PCR success. Designing primers with degenerate bases can help accommodate these variations.
Why is choosing the right rRNA gene target important for PCR?
16S rRNA targets prokaryotes, such as bacteria and archaea. For eukaryotes, like Vetigastropoda, 18S rRNA primers are more appropriate. Using the wrong target can result in no amplification or non-specific products.
Can low DNA concentrations still produce successful 16S PCR results?
Yes. DNA concentrations measured below 10 ng/µL by NanoDrop do not always prevent successful PCR or sequencing. Sample quality and PCR conditions also affect the outcome, so low readings should be interpreted cautiously.
How important is it to use positive controls in 16S rRNA PCR?
Very important. Testing primers on a positive control template confirms that they can amplify the target sequence. Without validation, failure in samples may reflect primer issues rather than sample quality.
What issues arise from crosstalk between 18S and ITS-2 regions in PCR?
Amplification can become non-specific if primers bind both 18S rRNA and ITS-2 regions. This crosstalk can cause mixed or misleading results, so primer design must minimize overlap with ITS-2 when targeting eukaryotic rRNA genes.


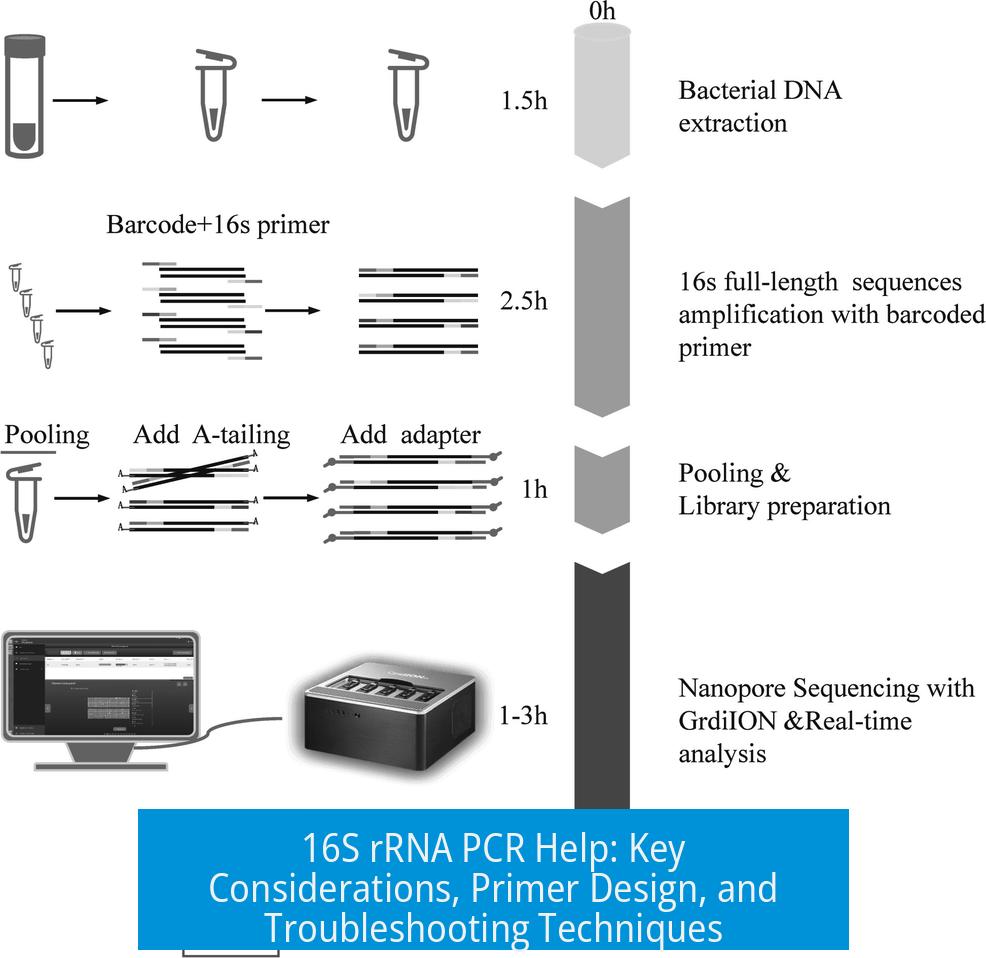

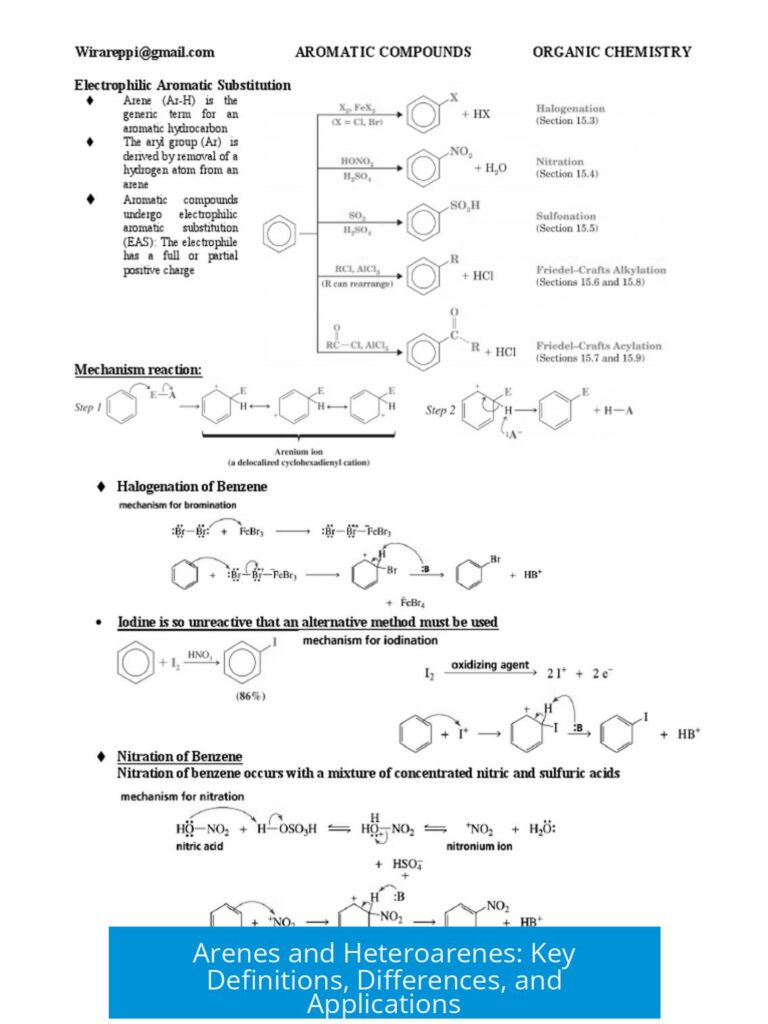
Leave a Comment