How to Generate 3D Structures from a Given Amino Acid Sequence

To generate a 3D structure (tertiary structure) from an amino acid sequence, one typically uses specialized computational tools that predict the protein’s folding based on known templates or de novo methods. These tools take the primary amino acid sequence as input and output a three-dimensional conformation. The choice of tool depends on the availability of structural templates similar to your protein of interest.
Understanding Protein Structures
- Primary structure: The linear sequence of amino acids.
- Secondary structure: Local folding patterns (e.g., alpha-helices, beta-sheets) often illustrated in 2D formats such as dot bracket notation.
- Tertiary structure: The complete 3D folding of a protein molecule, which determines its function.
The goal in generating 3D structures from sequences is to predict this tertiary structure accurately.
Using SWISS-MODEL for Template-Based 3D Structure Prediction
SWISS-MODEL (accessible at swissmodel.expasy.org) is the most straightforward tool for predicting tertiary structures when similar protein structures exist in databases.
- Submit the amino acid sequence to the web server.
- The server searches for homologous known structures (templates).
- It builds a 3D model based on these templates.
This approach relies on evolutionary similarity. If a close enough template does not exist, the model quality may be poor or unavailable.
Predicting Structure without Templates: Robetta
In cases where no similar protein structure is known, Robetta offers a solution. It uses de novo or hybrid modeling methods to attempt folding predictions.
- Robetta generates models without needing known templates.
- This is essential for novel or unique proteins.
- However, predictions may be less accurate compared to template-based methods.
Other Tools and Visualization Platforms
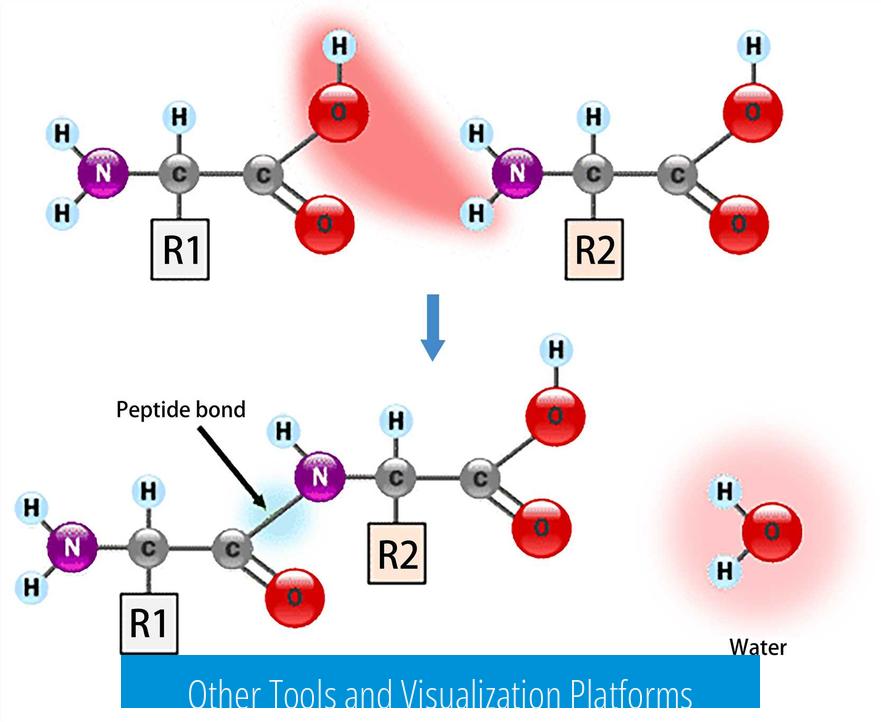
Secondary structure tools like NUPACK, Vienna2, and UnaFold provide predictions of local folding but do not generate full 3D structures. They can help visualize intermediate folding patterns.
The Eterna project uses innovative gamification to support tertiary structure modeling and visualization. It offers an interactive platform based on scientific puzzles to enhance understanding of protein folding.
Summary of Approaches
| Tool | Approach | Best For |
|---|---|---|
| SWISS-MODEL | Template-based homology modeling | Proteins with known similar structures |
| Robetta | De novo or hybrid modeling | Novel proteins lacking structural templates |
| NUPACK/Vienna2/UnaFold | Secondary structure prediction | Local folding visualization, not 3D modeling |
| Eterna Project | Tertiary structure visualization and gamification | Interactive learning and structure manipulation |
Key Takeaways
- Start with the amino acid (primary) sequence for 3D modeling.
- SWISS-MODEL is best if a similar protein structure is available.
- Use Robetta when no templates exist for your sequence.
- Secondary structure tools aid in understanding local folding but don’t provide full 3D structures.
- Exploring platforms like the Eterna project can enhance visualization and engagement with protein structures.
How to Generate 3D Structures from a Given Amino Acid Sequence
Wondering how to peek into the physical shape of your protein from just a string of amino acids? Generating a 3D structure from a given amino acid sequence boils down to using specialized computational tools that predict how your protein folds into its functional form. The key step is moving from the linear amino acid list—the primary structure—to the complex, spatial 3D shape: the tertiary structure. Let’s walk through the essentials and the tools you’ll want to put in your bioinformatics toolbox.
Conceptually, proteins start as simple lines—the amino acid sequence. This is called the primary structure. But proteins don’t stay linear; they twist and fold into shapes, first forming local patterns known as secondary structures (think alpha-helices and beta-sheets). However, the magic lies in the tertiary structure, the full 3D twist and turn of the entire protein that determines its function.
From Primary to 3D: What You Actually Want

Many people mix up the terms and settle for secondary structure predictions—simple 2D diagrams that show local folding patterns. These are neat, often depicted in dot bracket notation, but they are just the appetizer. The main course is the tertiary structure—the 3D protein model vital for understanding function, drug binding, and interactions.
So your quest starts with that linear sequence of amino acids—the primary structure. The goal? Generate a realistic 3D model. But how?
The Swiss SWISS-MODEL: Your First Port of Call
For many, the easiest and fastest route is SWISS-MODEL. Submit your amino acid sequence here, and this web server searches for similar structures in databases. If your protein resembles anything already catalogued, SWISS-MODEL stitches together a 3D model for you.
- Upload your sequence, follow the simple prompts.
- The server scans for matching templates—that is, known 3D structures with sequences close to yours.
- It builds a predicted tertiary structure based on these templates.
Remember: SWISS-MODEL depends heavily on the existence of similar proteins already solved. If your protein is well-studied or falls within a known family, this will likely work like a charm.
Oops, No Template? Meet Robetta
But what if your protein is a rebel—unique, novel, or simply not in the lucky club of known structures? That’s where Robetta enters the scene. This tool is designed specifically for de novo or hybrid modeling, which means it can predict structures without a close template to copy.
- Robetta uses machine learning and physical principles to fold your protein from scratch.
- It can be slower and less certain than template-based methods but is invaluable for unknown proteins.
- Great for novel proteins, engineered sequences, or those alien proteins scientists joke about.
So when SWISS-MODEL throws its hands up, give Robetta a shot. It’s like the improvisational actor of protein folding.
Secondary Structures: Helpful Sidekicks
Sometimes, you may want to peek at intermediate folding steps without diving straight into full 3D models. There are handy tools—like NUPACK, Vienna2, and UnaFold—which excel at predicting and visualizing secondary structures.
Think of these as handy sketches showing local twists and loops. While not the grand finale, these predictions help you understand folding patterns or refine your approach for more detailed 3D modeling.
Visualizing Your Creation: The Eterna Project
Got a 3D structure predicted? Now you want to see it in all its glory. Enter the Eterna Project powered by Stanford. This platform offers interactive visualization tools for tertiary structures wrapped in a gamified interface. It’s a fun, engaging place to explore how your protein folds and behaves.
- Interactive: manipulate and examine structures like a 3D puzzle.
- Connects community science and research.
- Publishes crowd-sourced design solutions, adding authenticity.
In short, Eterna adds a dash of playfulness to the usually math-heavy world of protein folding.
Putting It Together: A Simple Roadmap
- Start with your amino acid sequence (primary structure).
- Try SWISS-MODEL for template-based 3D predictions. It’s quick and accurate if similar structures exist.
- If no template is found, switch to Robetta for de novo modeling.
- Use secondary structure tools like NUPACK or Vienna2 if you want to understand folding patterns before or alongside 3D prediction.
- Visualize your models through the Eterna Project or similar visualization software.
Why Should You Care? Real Benefits at Your Fingertips
Knowing a protein’s 3D structure isn’t just academic trivia—it’s crucial for drug design, understanding diseases, and bioengineering new proteins. Rather than relying on pricey and time-consuming lab methods like X-ray crystallography, computational predictions open the door for rapid insights.
Plus, with tools constantly improving, even a novice can upload a sequence and get a predicted shape in minutes. This democratizes structural biology, getting everyone from students to startups in on the fun.
Trusting the Predictions: How Reliable Are They?
Template-based models like those from SWISS-MODEL are generally reliable, especially with high-quality templates. De novo tools like Robetta provide good starting points but should be experimentally validated when possible. Keep in mind, no prediction is perfect; proteins fold dynamically, and computational models are just snapshots.
If you want to check your predicted structure’s quality, many servers report a confidence score—don’t skip that! It helps decide if your model is ready for prime time or needs more work.
Final Tip: Always Validate and Iterate
Don’t stop at generating a model. Use visualization to inspect it for clashing side chains or weird folds. Iterate if needed. You might try running the model through molecular dynamics simulations or seek expert advice.
Generating 3D structures from amino acid sequences is no longer the domain of only specialized labs. With tools like SWISS-MODEL, Robetta, and visualization platforms like Eterna, anyone with a sequence can glimpse their protein’s shape. Ready to give it a shot? Your protein’s 3D journey awaits!
What is the difference between primary, secondary, and tertiary protein structures?
Primary structure is the linear amino acid sequence. Secondary structure shows local folds like alpha helices or beta sheets in 2D. Tertiary structure is the full 3D shape of the protein, which most prediction tools aim to generate.
How does SWISS-MODEL generate a 3D structure from an amino acid sequence?
SWISS-MODEL uses your amino acid sequence to find similar known protein structures as templates. It builds the 3D model by aligning your sequence with these templates. The accuracy depends on the availability of related structures.
Which tool should I use if no template structure exists for my protein?
Use Robetta for proteins without known templates. It can create 3D structures from scratch or by combining modeling methods. This helps generate models for novel proteins lacking similar known structures.
Can I visualize secondary structure predictions before generating the full 3D model?
Yes. Tools like NUPACK, Vienna2, and UnaFold provide secondary structure predictions. They offer 2D visualizations which can be useful for understanding local folding before tertiary structure modeling.
How can I visualize or interact with tertiary protein structures online?
The Eterna project offers interactive tools for visualizing tertiary structures. It uses gamification to help explore protein folding and allows users to engage with 3D models within their platform.



Leave a Comment