Blunt End Ligation: Overview, Challenges, and Alternatives
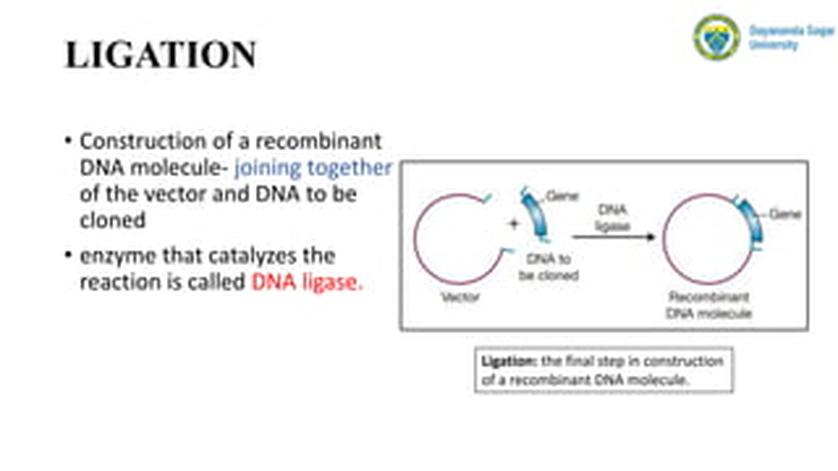
Blunt end ligation is a molecular cloning technique that joins DNA fragments without overhanging nucleotides, but it is generally inefficient compared to sticky end ligation methods. Due to its low efficiency and practical limitations, blunt end ligation is rarely the preferred method for cloning. Alternative methods such as Gibson assembly have effectively replaced blunt end ligation in many applications due to higher efficiency and better control over insert orientation.
Understanding Blunt End Ligation
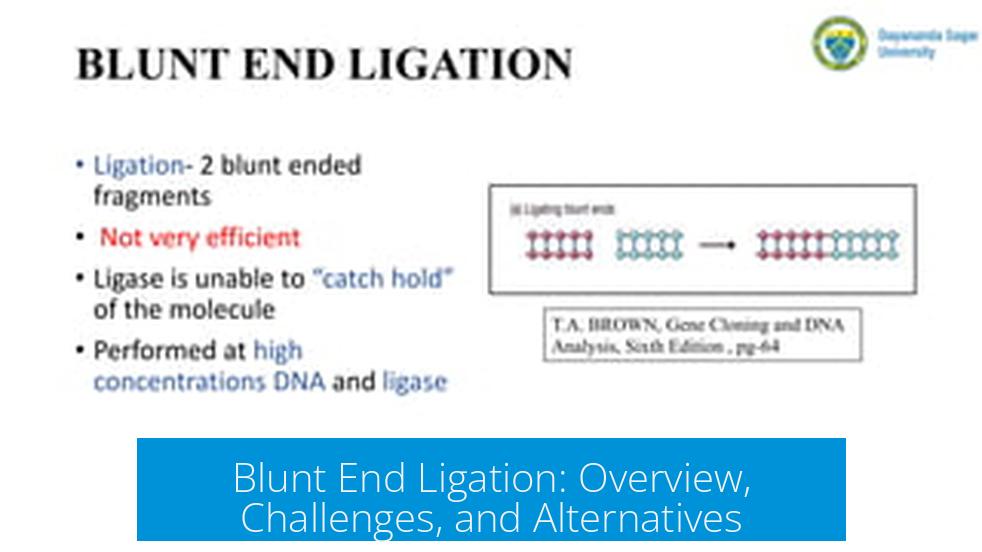
Blunt end ligation involves joining two DNA fragments whose ends are flush with no overhanging nucleotides. The process uses DNA ligase to covalently link the 3′ hydroxyl and 5′ phosphate groups at these blunt ends.
- Blunt ends lack single-stranded overhangs that facilitate base pairing.
- Ligation depends solely on the ligase enzyme for joining DNA strands.
- Commonly produced by restriction enzymes that cleave both strands symmetrically.
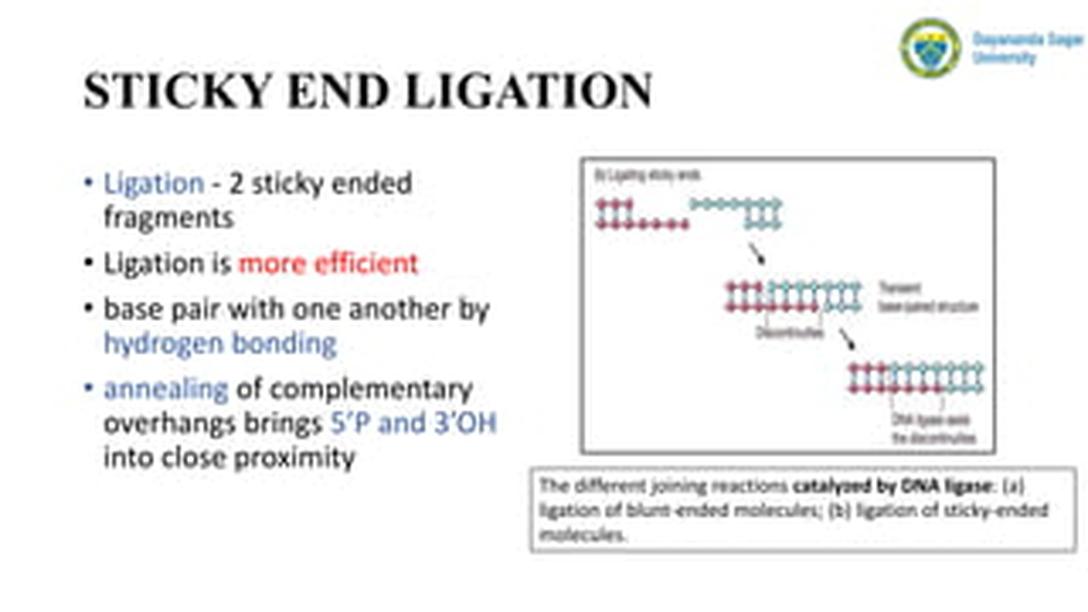
This contrasts with sticky end ligation, where complementary overhangs stabilize the interaction prior to ligase action, enhancing ligation efficiency.
Efficiency and Practicality of Blunt End Ligation
Blunt end ligation exhibits very low efficiency. The absence of complementary overhangs reduces the stability of fragment association and, consequently, reduces ligase access and activity.
Researchers generally advise against using blunt end ligation for routine cloning:
- Ligation occurs with lower yield compared to sticky ends.
- Increased chances of inserting the DNA fragment in the wrong orientation.
- Restriction sites can be lost when blunt ends are generated from sticky ones by end polishing.
These factors often lead to wasted time and increased experimental failure rates. One user noted blunt ligations often fail because “you end up going in backwards half the time” and compromise restriction sites.
Optimizing Blunt End Ligation

Despite its inefficiency, researchers sometimes optimize blunt end ligation with careful protocol tweaks. Successful blunt end ligation depends on multiple factors:
Vector to Insert Ratios
- Use increased molar ratios of insert relative to vector (e.g., 0.12 to 0.18 pmol of insert versus 0.02 pmol of vector).
- Higher insert concentrations favor collision and ligation events.
Incubation Time and Temperature
- Longer incubation periods improve ligation yield. Overnight incubation is recommended.
- Room temperature incubation is often preferable for blunt ends (around 20–25°C).
Ligase and Additives

- Use an ample amount of high-quality T4 DNA ligase (about 1 μL per 20–30 μL reaction).
- Adding poly(ethylene glycol) (PEG 8000) may enhance ligation by molecular crowding.
- Beware of PEG’s inhibitory effect on bacterial transformation if the ligation exceeds 1 hour.
Buffer and Handling
- Ensure the ligase buffer is fresh and has not undergone multiple freeze-thaw cycles.
- Aliquot the ligase buffer to reduce freeze-thaw damage.
Dephosphorylation
- Dephosphorylate the vector with calf intestinal phosphatase (CIP) to reduce self-ligation.
- This prevents vector recircularization without insert incorporation.
Bacterial Strain and Transformation
- Use highly competent strains like STBL2 or electrocompetent E. coli for better transformation efficiency.
- Blunt end cloning can be more challenging with large plasmids and inserts.
Adhering to these guidelines can modestly improve blunt end ligation success, but the process remains less reliable and slower than alternative methods.
Alternatives to Blunt End Ligation
Due to the inherent limitations of blunt end ligation, molecular biologists often prefer alternative cloning strategies.
Gibson Assembly: A Superior Method
Gibson assembly combines DNA fragments with overlapping ends in a single reaction facilitated by exonuclease, polymerase, and ligase activities.
- Creates precise, seamless fusions by generating complementary sticky ends in situ.
- Ensures correct insert orientation via unique overlapping sequences.
- Supports multiple DNA fragment assembly simultaneously.
Unlike blunt ligation, Gibson assembly uses overlap homology to guide correct fragment joining, eliminating orientation errors.
Use of Bridging Oligos in Gibson Assembly

When the insert and vector lack homology, single-stranded bridging oligos can be designed.
- Oligos provide overlapping sequences to facilitate assembly.
- Oligos can also introduce new restriction sites for downstream applications.
This approach offers flexibility without needing to modify the original insert or vector sequences extensively.
NEBuilder HiFi Assembly
NEB offers a commercial alternative to traditional Gibson assembly called NEBuilder HiFi DNA Assembly.
- Reportedly achieves higher efficiency and fewer errors.
- Compatible with complex multi-fragment assemblies.
- Includes detailed protocols on the NEB website for optimized results.
Comparing Blunt End Ligation and Gibson Assembly
| Feature | Blunt End Ligation | Gibson Assembly |
|---|---|---|
| Efficiency | Low; unreliable especially for large fragments | High; efficient even with multiple fragments |
| Insert Orientation | Random; often backward insertion occurs | Directed; overlaps ensure correct orientation |
| Requirement for Overhangs | None; blunt ends only | Requires overlap sequences (can be added via oligos) |
| Time Required | Long ligation times (overnight or longer) | Short reaction time (~15-60 minutes) |
| Restriction Site Preservation | Often lost through blunting | Maintained or engineered via oligos |
| Transformation Efficiency | Lower due to ligation inefficiency | Higher; clean reaction mixtures |
Conclusion and Recommendations
- Blunt end ligation remains a valid but inefficient cloning method.
- Optimize vector-to-insert ratios, incubation times, and ligase concentrations to improve blunt ligation success.
- Use dephosphorylation and suitable bacterial strains to enhance cloning outcomes.
- Whenever possible, use Gibson assembly or NEBuilder HiFi assembly for higher efficiency and orientation control.
- Incorporate bridging oligonucleotides in Gibson assembly when inserts and vectors lack homology.
- Consider alternatives first before opting for blunt end ligation in cloning strategies.
Blunt End Ligation: Why It’s the Underdog and When to Rethink Your Strategy
Ever tried blunt end ligation and felt like you’re pouring your time into a black hole? It’s a classic problem for many molecular biologists. Blunt end ligation is generally low in efficiency and often less practical compared to sticky end ligation or modern cloning techniques. Let’s unpack why it’s often left at the bottom of the toolbox, how to possibly optimize it, and when to drop it for better alternatives.
Why Blunt End Ligation Has a Reputation for Frustration
Here’s the blunt truth: ligating DNA strands that don’t have sticky overhangs results in poor ligation efficiency. Double blunt ends are just very low efficiency, with lots of failed ligations waiting to happen. People often say, “don’t waste your time on blunt end cloning.”
One huge drawback is the nonspecific nature of blunt ends — they can ligate in either orientation. This means half the effort can be wasted on “backwards” inserts. Add to this the fact that blunting sticky ends destroys useful restriction sites, and it becomes clear why many researchers say blunt ligation should be avoided for simple cloning projects.
It’s like using a spoon to cut steak—technically possible, but there are better tools.
How to Squeeze More Out of Blunt End Ligation
If you must, there are ways to improve blunt end ligation outcomes:
- Manipulate vector-to-insert ratios. Higher insert ratios—like 0.12 to 0.18 pmol—can push the odds in your favor.
- Incubate longer, overnight at room temperature, or even over a weekend if you’re desperate.
- Use ~0.02 pmol of vector, ~0.06 pmol of insert, and 1μL of standard NEB ligase in 20–30 μL reaction volumes.
- Add PEG 8000 to the mix to facilitate molecular crowding, though beware NEB’s advice to not extend reaction times beyond an hour with PEG present, as it can hurt transformation.
- Don’t reuse or over-thaw your ligase buffer—aliquoting can save your day.
- Experiment with adding more ligase, or switch to high-concentration ligase strains available at NEB.
- Consider dephosphorylating your vector using calf intestinal phosphatase (CIP) to prevent vector self-ligation, improving insert incorporation chances.
- Choose your bacterial host carefully. High-efficiency electrocompetent E. coli strains, like STBL2, can make a meaningful difference.
Does any of this sound like a tedious series of “maybes”? That’s because blunt ligation demands patience and precision, with no guarantees. You might get lucky, but often, luck feels insufficient for scientific reliability.
Why You Might Want to Say Goodbye to Blunt End Ligation
Is blunt end ligation ever truly advisable? It’s seldom ideal. Why settle for a method that wastes valuable time, ruins restriction sites, and doesn’t guarantee right orientation of your insert?
Cloning is all about efficiency and predictability. The last thing you want is a reaction that treats your DNA like a jigsaw puzzle tossed in the air. Sticky end ligation and, even better, modern assembly techniques, often save the day.
Modern Alternatives: Gibson Assembly and NEB HiFi — The New Kids on the Block
Meet Gibson Assembly, the cloning wizardry that magic users envy. Using a mix of exonucleases, polymerases, and ligases, it engineers overlapping sticky ends for you. This ensures the insert locks into the vector in the correct orientation every single time.
One great tip: you can even use bridging single-stranded DNA oligos to create homology between your vector and insert if they otherwise have none. That’s like adding a customized handshake between your DNA fragments, making sure they’re introduced correctly.
Researchers rave about using NEBuilder HiFi mix (an improved version of Gibson Assembly from NEB). It works well for tricky assemblies and lets you design your insert with added restriction sites, enabling future modifications easily.
For example, you can add unique restriction sites right in the bridging oligo sequence, giving you the power to excise or swap inserts later with surgical precision.
Thinking Beyond Cloning: Making Smart Choices
So, should you relegate blunt ligation to history? Almost always, yes. Unless it’s your last resort, alternatives like Gibson Assembly offer better success rates, ensure correct insert orientation, and preserve your precious restriction sites.
Are you stuck with a blunt-end project? Try optimizing with some of the tips above. But, if you want the cloning magic that doesn’t feel like rolling dice every time, explore the Gibson route.
Ultimately, cloning success demands efficiency, precision, and reproducibility—qualities that blunt end ligation rarely guarantees.
Wrapping It Up
Blunt end ligation is a tricky and inefficient DNA joining method that often leads to frustratingly low success rates. While it can be optimized by adjusting vector-to-insert ratios, incubation conditions, ligase amount, and bacterial strains, modern molecular biology techniques like Gibson Assembly provide better reliability and convenience.
Switching to assembly techniques is not just about embracing new tech—it’s about respecting your time and research integrity.
So next time you’re planning your cloning strategy, ask yourself: Why settle for less? Give your experiment the edge it deserves and say goodbye to blunt end ligation woes!
And hey, if you do decide to wrestle with blunt ligations, don’t forget to bring your patience, some PEG, and maybe a cup of strong coffee.
What causes the low efficiency of blunt end ligation?
Blunt end ligation lacks sticky overhangs, reducing base pairing between insert and vector. This lowers ligation efficiency and makes cloning less reliable. Many prefer other cloning methods to avoid wasted time.
How can I optimize a blunt end ligation reaction?
- Use higher insert to vector ratios, around 0.12 to 0.18 pmol of insert.
- Incubate ligation overnight at room temperature or longer if needed.
- Try more ligase or add PEG 8000, but limit reaction time if PEG is used.
- Use fresh ligase buffer and high-efficiency bacteria like STBL2 or electrocompetent cells.
Why should I consider alternatives like Gibson assembly instead of blunt ligation?
Gibson assembly creates sticky ends via nuclease and ligates inserts reliably in the correct orientation. It is more efficient and flexible, allowing use of bridging oligos to join fragments with no homology.
Can blunt end ligation be improved by using dephosphorylation?
Dephosphorylation (e.g., using CIP) can prevent vector self-ligation but doesn’t increase blunt end ligation efficiency much. It’s one of several strategies but not a solution to low success rates alone.
What vector to insert ratios are recommended for blunt ligation?
Higher insert:vector ratios help. Use about 0.02 pmol of vector and 0.06 pmol or more of insert in a 20-30 µL reaction volume. Adjust ratios experimentally for best results.
How does incubation temperature affect blunt end ligation?
Blunt ligations usually work best at room temperature with long incubation times, sometimes overnight or over a weekend. Unlike sticky end ligations done in the fridge, warmth helps ligase efficiency here.




Leave a Comment