Understanding DNA Fingerprinting
DNA fingerprinting, also called DNA profiling, identifies individuals by analyzing unique patterns in their DNA. It plays a vital role in forensic investigations and paternity testing. The method relies on detecting variations in DNA sequences that differ from person to person.
Core Techniques in DNA Fingerprinting
- Restriction Enzyme Digestion: These enzymes cut DNA at specific sequences, producing fragments of varying lengths. This step is essential because it prepares the DNA sample for analysis by generating reproducible fragment patterns.
- Gel Electrophoresis: DNA fragments are placed in an agarose gel and exposed to an electrical field. Smaller fragments move faster through the gel, resulting in size-based separation. This process produces a distinct band pattern.
- Visualization of DNA Bands: After electrophoresis, the fragments are stained and observed as bands under UV light. Each band pattern forms the individual’s unique DNA fingerprint.
Key Genetic Markers Used
Two main types of DNA markers provide the basis for fingerprinting:

- Variable Number Tandem Repeats (VNTRs): These are long sequences repeated multiple times at specific loci. Different individuals have different repeat numbers. VNTR analysis is characteristic of classical DNA fingerprinting.
- Short Tandem Repeats (STRs): These are shorter repeated sequences. STRs show high variability in populations and are now favored in modern DNA profiling due to their sensitivity and speed.
Historical and Modern Approaches
Originally, forensic work focused on VNTRs combined with restriction enzyme digestion, producing band patterns on gels. Current methods mainly utilize STR analysis, employing polymerase chain reaction (PCR) amplification for precise and rapid detection. The shift enhances reliability and requires less DNA.

Summary of DNA Fingerprinting Process
| Step | Description |
|---|---|
| DNA Extraction | Isolate DNA from cells of the individual. |
| Restriction Enzyme Digestion | Cut DNA at specific sequences to generate fragments. |
| Gel Electrophoresis | Separate fragments by size in a gel matrix. |
| Visualization | Stain DNA to reveal unique band patterns. |
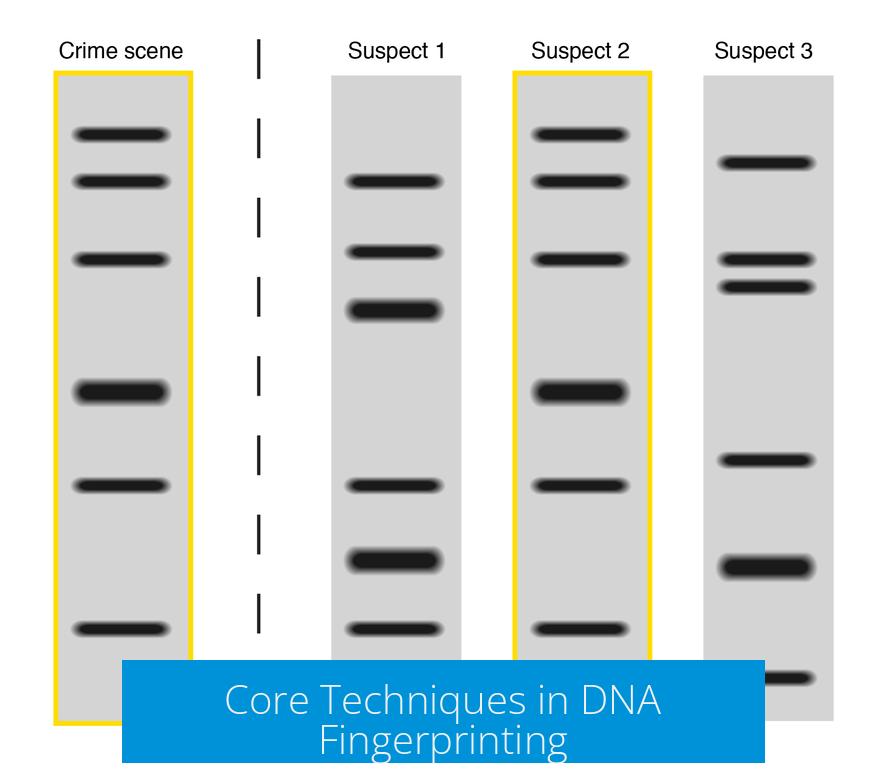
| Pattern Analysis | Compare bands to identify genetic matches. |
Further Learning
Scientific reviews on platforms such as PubMed provide comprehensive insights into restriction enzymes and DNA marker analysis. Visual aids and tutorials available on YouTube can assist in grasping the techniques involved.
- DNA fingerprinting detects unique repeat sequences in DNA.
- Restriction enzymes and gel electrophoresis enable fragment separation.
- VNTRs were used historically; STRs dominate modern profiling.
- The process is crucial for forensics and paternity verification.
DNA Fingerprinting: Cutting, Running, and Revealing Your Genetic Signature
DNA fingerprinting is a powerful technique that creates a unique genetic profile for an individual by analyzing specific regions of their DNA. Often dubbed DNA profiling, it has revolutionized fields like forensics and paternity testing by offering a precise and reliable method of identification. But how exactly does this molecular magic work? Let’s unravel the science behind these viral rungs on the ladder of justice and family drama.
Imagine being able to read someone’s personal bar code, not on a product but on a microscopic thread inside their cells. DNA fingerprinting achieves exactly that by focusing on particular DNA sequences that vary highly between people. This uniqueness serves as a tool to answer questions like: Who left that fingerprint at the crime scene? Or, is Daddy really who you think he is?
The Building Blocks: Restriction Enzymes and Cutting DNA
At the heart of classical DNA fingerprinting lie restriction enzymes—tiny molecular scissors with a sharp taste for specific DNA sequences. These enzymes cut DNA at predetermined sites, chopping the long DNA strands into fragments of various lengths. This process, called restriction enzyme digestion, is crucial because it generates the pieces you’ll later sort and analyze.
Think of it as shredding a book by cutting only at certain words, so the resulting paper strips vary by length and content. The pattern of these strips will be unique to the original book edition—just like DNA fragments differ among individuals.
Separating the Fragments: Gel Electrophoresis
Once the DNA gets sliced, how do you make sense of the jumble? This is where gel electrophoresis comes in. You place the DNA fragments into a gel matrix and apply an electric current. Because DNA molecules have a negative charge, they swim toward the positive electrode.
The magic happens in how fast they move. Smaller fragments slip through the gel pores more quickly, while larger ones lag behind. After some time, this electrical race produces a collection of distinct DNA bands aligned by size. It’s like a molecular marathon displaying runners sorted by their speed.
Seeing the Patterns: Visualization of DNA Bands
To see these DNA fragments, scientists stain the gel with dyes that make the bands visible under UV light. What appears is a series of distinct bands, some thick, some thin, scattered along the gel. Each person’s band pattern forms a unique fingerprint. No two individuals (except identical twins) share the same arrangement, providing the molecular basis of identification.
Variable Regions: VNTRs and STRs—Nature’s Barcode
What pieces of the genome do fingerprinting methods examine? Enter Variable Number Tandem Repeats (VNTRs) and Short Tandem Repeats (STRs), the real stars behind the curtain.
- VNTRs are longer DNA segments with sequences repeated several times in a row. The number of repeats varies significantly between people, which makes VNTRs very polymorphic—perfect for classical fingerprinting.
- STRs are the smaller siblings: shorter repeat sequences that modern DNA profiling targets. They allow faster and more precise analysis, even with tiny or degraded samples.
Both VNTRs and STRs serve as unique, measurable landmarks in genomes. If you think examining your street address is unique, then comparing someone’s VNTRs or STRs is like having a genetic address you can’t fake.
Old School vs. New School DNA Fingerprinting
Here’s a quick flashback: DNA fingerprinting was once a heavier, trickier work involving VNTR analysis and restriction enzymes. Early methods took longer and required high-quality DNA, limiting their use.
Modern methods have refined this process tremendously by focusing on STRs and automated analysis. These advances speed up results and increase accuracy. It’s like switching from dial-up Internet to fiber optic — the difference is dramatic.
Why Should You Care About DNA Fingerprinting?
Perhaps you’ve seen on TV how DNA evidence “cracks the case.” While dramatized, there’s real science behind it. DNA fingerprinting has:
- Resolved criminal investigations by matching suspects to crime scene samples.
- Settled paternity and maternity questions with indisputable genetic proof.
- Helped in missing person identification and disaster victim recovery.
- Simplified biodiversity studies and wildlife tracking.
If you’re curious how DNA profiling works practically, there are many educational videos on YouTube explaining the steps visually. For deep dives and scientific data, PubMed reviews provide detailed explorations into restriction enzyme digestion and VNTR/STR analysis.
Tips When Exploring DNA Fingerprinting Yourself
- Understand the basics: Grasp how enzymes cut DNA and how electrophoresis separates fragments first. This foundation helps comprehend the bigger picture.
- Seek credible sources: Reliable websites and scientific reviews avoid oversimplifications or false promises about DNA test results.
- Compare methods: Know whether the test you’re considering uses classical VNTR analysis or modern STR-based profiling.
Even if you’re not planning to work in a lab, DNA fingerprinting’s applications touch many aspects of life—medicine, law, genealogy, and more. It’s a modern marvel that uncovers truths locked in our very cells.
Final Thought
Ever wonder why your DNA is like a secret signature nobody else shares? DNA fingerprinting reveals this biological identity with stunning precision. It is a remarkable tool blending molecular biology and technology to solve mysteries, settle disputes, and unveil connections we wouldn’t see otherwise.
So next time you hear “DNA fingerprinting,” picture those tiny molecular scissors, electric gels, and glowing bands—proof that science can illuminate invisible threads tying our stories together.
For more details, check out the DNA profiling Wikipedia page to explore its history and development. Curious minds, grab your popcorn and dive into DNA mysteries!
What role do restriction enzymes play in DNA fingerprinting?
Restriction enzymes cut DNA at specific sequences. This creates fragments needed for analysis. These cuts allow unique patterns to form during fingerprinting.
How does gel electrophoresis separate DNA fragments?
DNA fragments are run through a gel using electricity. Smaller fragments move faster. This separates fragments by size, forming distinct bands.
What makes VNTRs useful for DNA fingerprinting?
VNTRs are DNA regions with repeating sequences. The number of repeats varies between people. This variation helps distinguish individual DNA profiles.
How do STRs differ from VNTRs in DNA profiling?
STRs are shorter repeat sequences. Modern profiling mainly uses STRs because they are easier to analyze. VNTRs were used in older methods.
Why is visualization of DNA bands important?
Visualization shows the separated DNA fragments as bands. These bands form a unique pattern for each person. This pattern is the DNA fingerprint.





Leave a Comment