Protocol and Excel Template for Mitochondrial Copy Number Analysis via qPCR
The mitochondrial copy number (mtDNA copy number) can be calculated using the formula 2 × 2^dCt, where dCt is the difference between the Ct values of a nuclear gene and a mitochondrial gene (dCt = Ctnuclear – CtmtDNA).
1. Sample Preparation

For mtDNA copy number analysis, reverse transcription (RT) is unnecessary since RT is used for RNA studies. Extract genomic DNA directly from your samples. Dilute the DNA appropriately to ensure consistent qPCR reactions. This simplifies the preparation step, focusing only on DNA quality and concentration.
2. qPCR Protocol
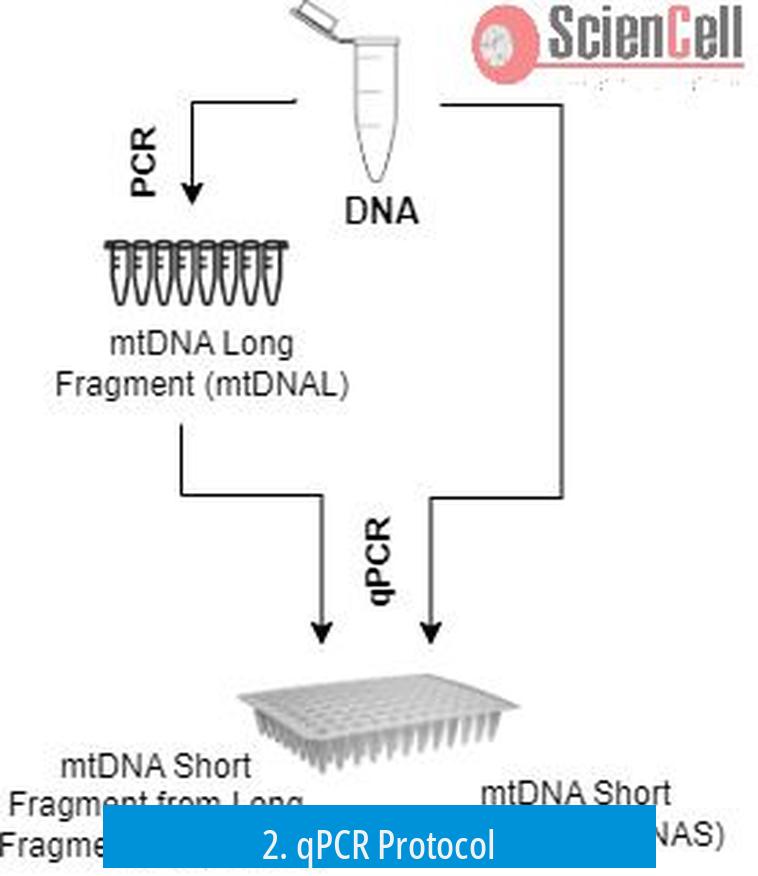
- Use a multiplex TaqMan probe assay that targets mtDNA and a nuclear reference gene in the same reaction.
- Design primers and probes tailored for your experimental model. Published literature often provides validated primer sets.
- Run the qPCR reaction according to standard cycling conditions suitable for your probes and primers.
- Obtain Ct values for both mitochondrial and nuclear genes for each sample.
3. Calculation of Mitochondrial Copy Number
Calculate the difference in Ct (dCt) by subtracting the mitochondrial gene Ct from the nuclear gene Ct. Apply the formula:
| Step | Calculation |
|---|---|
| dCt | Ctnuclear – CtmtDNA |
| Average copy number per cell | 2 × 2dCt |
This result estimates the average mitochondrial DNA copy number per cell, assuming two copies of the nuclear gene per diploid genome.
4. Using Excel for Data Analysis
Creating a dedicated Excel template is optional. Since the calculation is straightforward, manually inputting Ct values and formulas in Excel is preferable. This allows visualization of each step and helps identify possible errors or inconsistencies in your data.
Set up columns for sample ID, nuclear Ct, mitochondrial Ct, dCt, and copy number. Use simple cell formulas to automate the calculation while maintaining transparency.
Key Takeaways
- Mitochondrial copy number uses the formula: 2 × 2(Ctnuclear – CtmtDNA).
- Extract DNA directly; no reverse transcription required.
- Utilize a multiplex TaqMan qPCR assay targeting both nuclear and mitochondrial genes.
- Choose primers and probes based on your specific model or published sequences.
- Manual calculation in Excel promotes understanding and troubleshooting.
Can Anyone Please Help Me with Protocol and Excel Template for Mitochondrial Copy Number Analysis via qPCR?
Here’s the bottom line: Mitochondrial copy number calculation via qPCR is straightforward. You extract your DNA, run a multiplex qPCR comparing your mitochondrial gene Ct and a nuclear gene Ct, and calculate copy number using the formula 2*2dCt, where dCt = Ctnuclear – Ctmitochondrial. Surprisingly, there’s no flashy Excel template you must use — it’s better to calculate yourself to catch any curveballs.
Now, if you came here hoping to grab a ready-made Excel file and a strict step-by-step guide, you might be a tiny bit disappointed. But stick with me, because I’ll walk you through the nitty-gritty details, share tips from real-world qPCR warriors, and explain why a DIY approach beats copy-pasting from a template every day.
What’s All the Fuss About Mitochondrial Copy Number via qPCR?
Mitochondrial DNA (mtDNA) copy number is often a biomarker of cellular health, energy metabolism, aging, and disease states. Scientists need reliable ways to measure how many mitochondria—or copies of mitochondrial genome—exist per cell. Quantitative PCR (qPCR) has earned its place as the gold standard for this, given its sensitivity and specificity.
But despite its obvious importance, the exact steps can feel murky. What genes to amplify? How to calculate the final values? And the dreaded question: “Excel template? Where is it?”
Step 1: Sample Prep — Simpler Than You Think
Forget about reverse transcription (RT). That’s for RNA, not DNA. Since we want mitochondrial DNA copy numbers, all you do is extract total DNA from your samples. Use your favorite DNA extraction kit or method. Then dilute the DNA to a concentration suitable for qPCR. This step is crucial because too much or too little DNA can skew results.
TIP: Run a pilot to optimize your DNA concentration. Some people overlook this and end up with inconsistent Ct values. Also, beware of contaminating nuclear mitochondrial DNA segments (NUMTs) – choosing specific primers helps minimize this.
Step 2: The Protocol — Multiplex TaqMan Probes Are Your Friends
There are many published protocols floating around. But a reliable and recommended strategy is a multiplex TaqMan probe assay that targets a mitochondrial gene and a nuclear gene simultaneously. This helps you get a direct comparison in the same reaction well, reducing variability.
You need to select primers and probes carefully. Papers in your specific model system often share validated sequences. If not, you might design new primers—just be sure to test their efficiency (should be roughly 90-110%) and specificity (no off-target amplification).
Here’s what a typical workflow looks like:
- Prepare qPCR mix with your DNA sample, primers, probes, and master mix.
- Run qPCR cycling conditions optimized for your primers.
- Obtain Ct values for your mitochondrial gene and nuclear gene.
The magic happens after.
Step 3: Calculating Mitochondrial Copy Number — No Black Magic, Just Math
You have these two Ct values: mitochondrial Ct and nuclear Ct. Subtract mitochondrial Ct from nuclear Ct to get dCt. Yes, nuclear Ct minus mitochondrial Ct is the official formula.
The formula to calculate average mitochondrial copies per cell is:
Average copy number = 2 * 2dCt
Why multiply by 2? Because every diploid cell has two copies of the nuclear gene (assuming your nuclear gene is single-copy). This formula gives you a relative—yet meaningful—estimate of mtDNA copies per cell.
Need an example? Let’s say nuclear gene Ct is 23 and mitochondrial gene Ct is 20.
- dCt = 23 – 20 = 3
- 2dCt = 23 = 8
- Average copy number = 2 * 8 = 16 copies per cell
Step 4: Excel Template — Or Just Your Calculator?
Despite the popularity of Excel in labs, professionals often find a template unnecessary—and dare I say, limiting. Calculating copy number yourself, either in a spreadsheet or even with a standard calculator, is far more insightful. You see each step, catch data entry errors early, and better understand your variations.
Sure, some may want a ready-made template, but trust a seasoned hand: building your own simple Excel sheet is empowering. It’s just subtraction, exponentiation, and multiplication. Plus, it’s easy to expand if you want to include replicate averages, normalization controls, or graphs.
Pro tip: Set up columns titled “Nuclear Ct”, “Mitochondrial Ct”, “dCt”, and “Copy Number”. Use Excel formulas like =B2-C2 for dCt and =2*POWER(2,D2) for copy number. Easy to audit, tweak, and share.
Why Trust This Method?
This approach is widely accepted and validated in the scientific literature. Using a multiplex qPCR with carefully designed primers, you minimize variability. Skipping reverse transcription removes complexity, focusing purely on DNA quant. The calculation formula is a neat application of qPCR principles in relative quantification.
It’s transparent—no opaque software hidden under the hood. Plus, it’s flexible. You can adapt it to different species or gene targets by changing primers.
Want to Make Your Data Rock?
- Run technical replicates (at least in triplicate) to reduce random variation.
- Include no-template controls to spot contamination early.
- Normalize your DNA input carefully.
- Validate primers for efficiency close to 100%.
- Stay consistent in sample handling.
Doing this means your calculated mitochondrial copy numbers reflect real biology, not lab chaos.
Final Thoughts
In short, mitochondrial copy number analysis via qPCR doesn’t need a complicated Excel template or a labyrinth of protocol steps. You just need:
- DNA extraction (skip RT)
- Multiplex qPCR with mitochondrial and nuclear gene primers/probes
- Ct measurement followed by the calculation using 2 * 2(nuclear Ct – mitochondrial Ct)
- A simple personal Excel or even calculator-based sheet for clarity
By mastering these basics, you turn a confusing topic into a straightforward lab skill. And when someone asks for the “protocol and Excel template,” you’ll say, “No problem. Here’s the math, the method, and the know-how to rock your mitochondrial DNA results.”
Ready to give it a shot? Grab your DNA, design your primers, and may your Ct values always be consistent!
What is the formula to calculate mitochondrial DNA copy number from qPCR data?
Use 2*2^dCt to get the average copy number per cell. dCt is the difference between the nuclear gene Ct and the mtDNA gene Ct values.
Do I need to perform reverse transcription (RT) before qPCR for mtDNA copy number?
No, RT is for RNA analysis. For mtDNA copy number, just extract and dilute your DNA before running the qPCR assay.
What type of qPCR protocol is recommended for mitochondrial copy number analysis?
A multiplex TaqMan probe protocol is advised. It uses nuclear gene references for relative quantification, but primers should fit your specific model.
Is there a ready-made Excel template for mitochondrial copy number calculation?
An Excel template is not necessary. It’s simpler to do the calculations manually to better understand the data and catch any errors.
How do I select primers and probes for my mtDNA copy number assay?
Look at published papers for suitable primers and probes or design your own based on the model system you work with. This ensures specificity.




Leave a Comment