CRISPR Design Questions: Essential Guidance on Vector Choice, Markers, and Validation
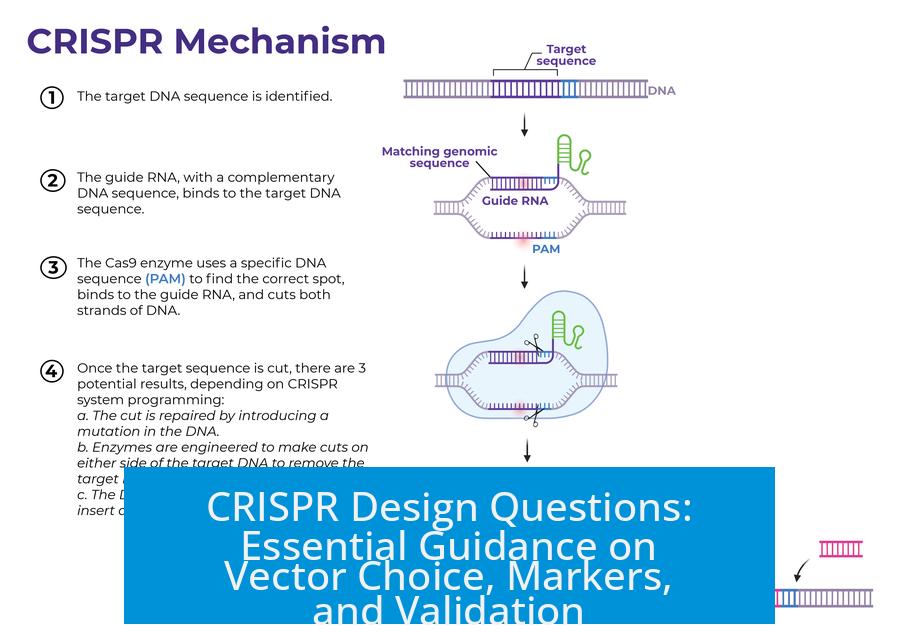
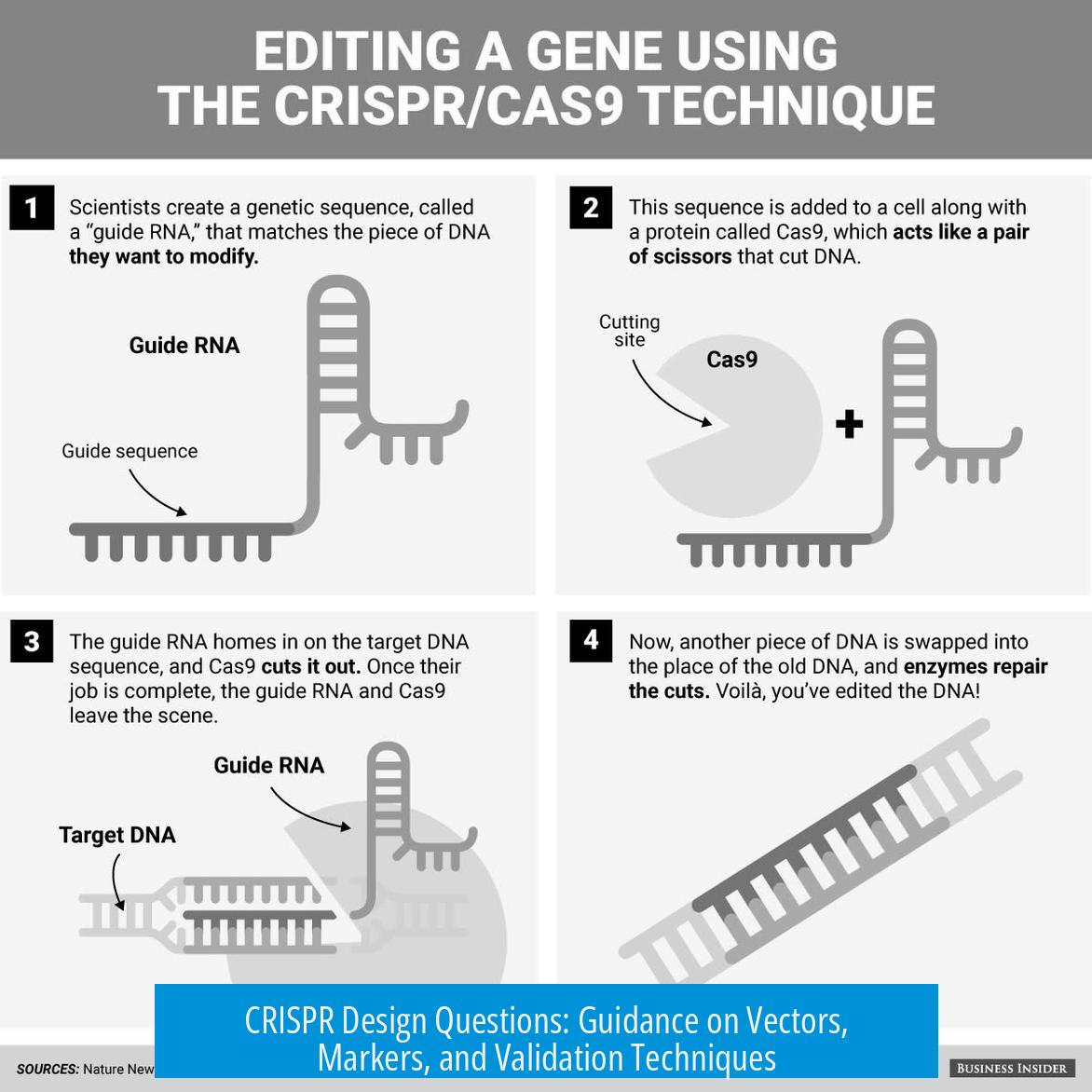
CRISPR design requires choosing suitable vectors, markers, and reliable validation methods. Vector choice impacts efficiency; combining fluorescent proteins (FP) with selection markers optimizes expression tracking. Confirming knockout (KO) success uses DNA and protein analyses to ensure precision. This article breaks down critical considerations for effective CRISPR experimental design.
1. Vector Choice and Delivery Methods
a) Lentivirus vs PiggyBac Recombinase
Lentiviral vectors remain common for stable gene integration, but alternatives exist. PiggyBac recombinase stands out as a simpler, more efficient option, yielding comparable stable insertion results. This approach facilitates easier cloning and reduces biosafety concerns linked with lentiviral packaging.
Compared to lentivirus, piggyBac enables transgene mobilization without viral production steps. This feature lowers time and resource demands, making it a preferred choice for researchers seeking efficiency.
b) Lentiviral Vector Packaging Choices
Users considering lentiviral systems must decide between self-packaging lentiviral particles or purchasing ready-made virus particles. Self-packaging allows customization but increases labor and requires biosafety facilities. Buying pre-packaged lentivirus simplifies workflows but may limit control over vector properties.
Clarifying this early is crucial. Vector performance and safety rely on packaging quality, so informed decisions affect transduction efficiency and experiment reproducibility.
2. Fluorescent Proteins and Selection Marker Strategies
a) Combining FP and Selection Marker for Cas9 Monitoring
Co-expressing Cas9 with a fluorescent protein and a selection marker optimizes tracking and enrichment of edited cells. The recommended design often uses a bicistronic cassette:
- Promoter – Cas9 (with NLS) – P2A – FP – polyadenylation signal
- New promoter – Selection marker – separate polyadenylation signal
This strategy ensures the FP only fluoresces if the entire Cas9 expression system is active, avoiding false positives. Using red or far-red FPs like mRuby or mKate2 reduces spectral overlap and background from commonly used green fluorescent proteins.
b) Selection Marker Choices and Optimization
Choosing an appropriate antibiotic resistance marker is cell-line dependent. Hygromycin is preferred in some contexts due to cell viability nuances, but puromycin and blasticidin remain popular. Performing a kill curve to establish minimal effective concentrations ensures specific selection without excessive toxicity.
c) Fluorescent Markers for gRNA Transfection Efficiency
Since gRNA plasmids are transiently expressed, green or blue fluorescent proteins serve best to monitor transfection efficiency quickly. The transient nature prevents prolonged marker expression, making these colors ideal for short-term tracking.
d) Fluorescence Intensity and Cell Type Considerations
Some cell types, such as mouse lines, show weak fluorescence post-transduction, reducing the utility of fluorescence-activated cell sorting (FACS). For cell lines like Jurkat, fluorescence sorting might also not improve outcomes if antibiotic selection is effective.
Using less common fluorescent proteins than GFP avoids confusion caused by widespread green fluorescence in plasmids. This precaution ensures clarity in multiplexed or long-term experiments.
3. Confirmation and Validation of Knockout
a) Validation Using ICE Analysis
Indel Correction Efficiency (ICE) analysis offers a quantitative method for assessing CRISPR-induced knockout efficiency. PCR amplifies the target genomic region followed by sequencing and computational analysis. This method estimates the fraction of alleles harboring disruptive mutations.
Choosing between using the heterogeneous KO pool or isolating single clones depends on the knockout percentage and research requirements. Clonal isolation enables precise downstream characterization but is labor-intensive. Pools provide a quicker, though less uniform, alternative.
b) Protein Level Assessment via Western Blot
Direct protein detection methods, such as western blotting, provide functional confirmation of the knockout effect. Measuring target protein reduction pre- and post-gRNA transfection reveals the phenotypic outcome more reliably than fluorescence intensity.
Sorting fluorescent cells alone only indirectly correlates with gene disruption, so confirming protein loss is crucial for accurate KO validation.
c) Considerations on Stable Cas9 Expression
Stable Cas9 expression introduces permanent genomic modifications, which may be unnecessary or undesirable. Alternative approaches include transient transfection of Cas9 with sgRNAs or cloning all gRNAs into a single Cas9/sgRNA backbone plasmid, allowing sequential or parallel testing without permanent changes.
This flexibility allows researchers to generate KO lines without enduring Cas9 integrations, simplifying interpretation and reducing potential off-target effects.
4. General Suggestions and Resources
a) Accessing Vectors from Public Repositories
Repositories like Addgene provide a broad selection of vetted lentiviral vectors for Cas9 and sgRNAs. Browsing such collections accelerates design by offering well-characterized constructs with community-validated performance data.
b) Tailoring Designs to Cell Type
Recognize the intrinsic properties of your target cells, including fluorescence capacity and antibiotic sensitivities. Adjust vector types, promoters, and markers accordingly. Pilot experiments are invaluable for defining optimum conditions specific to your system.
Summary of Key Points
- PiggyBac recombinase offers a more efficient alternative to lentiviral vectors for stable gene integration.
- Decide early whether to package lentivirus in-house or obtain ready-made viral particles based on lab capabilities.
- Cospacing Cas9 with red or far-red fluorescent proteins ensures fluorescence indicates full Cas9 expression.
- Use distinct promoters and polyadenylation signals for fluorescent markers and selection genes to avoid expression interference.
- Perform antibiotic kill curves to establish effective selection concentrations tailored to your cell line.
- Transient green or blue fluorescent proteins provide quick assessment of gRNA plasmid transfection efficiency.
- Fluorescence sorting may not improve outcomes in some cell types with weak fluorescence; selection markers can compensate.
- Validate knockouts by ICE sequence analysis and measure protein reduction by western blot for robust confirmation.
- Consider transient Cas9 expression methods to avoid permanent genomic modifications.
- Leverage Addgene and other repositories to source validated vectors and streamline experimental design.


Leave a Comment