GFP qPCR Primers: Essential Tools for Gene Expression Analysis
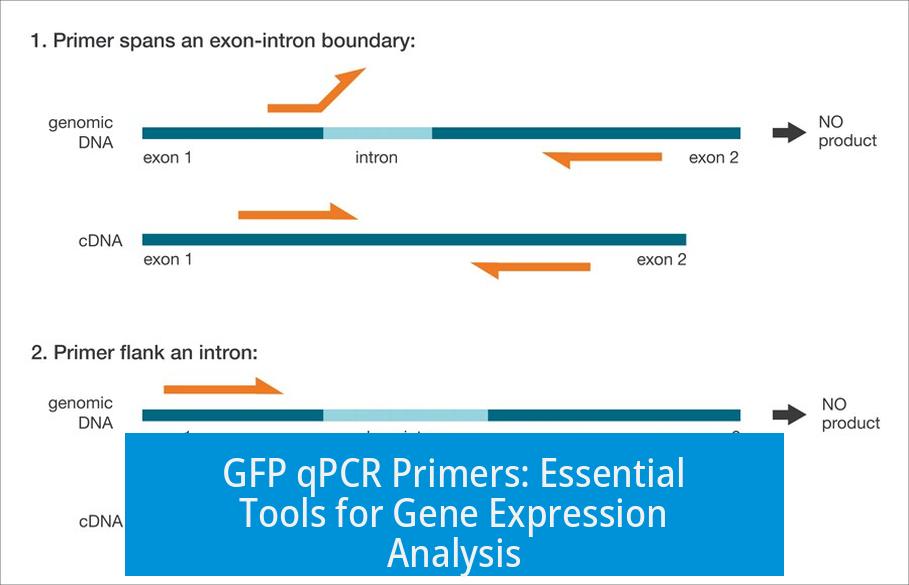
GFP qPCR primers are short sequences of nucleotides designed to specifically amplify the green fluorescent protein (GFP) gene during quantitative PCR (qPCR). These primers enable researchers to measure GFP expression levels accurately in various biological samples. GFP is widely used as a reporter gene in molecular biology studies to monitor gene expression and protein localization.
Understanding GFP and Its Importance in qPCR
GFP originates from the jellyfish Aequorea victoria and fluoresces green under UV or blue light. It serves as a marker to track gene expression, protein localization, or cellular processes. Quantitative PCR (qPCR) is a sensitive technique to quantify nucleic acids, making it ideal to measure GFP mRNA levels.
Design Principles for GFP qPCR Primers
- Specificity: Primers must target unique regions of the GFP gene to avoid amplifying non-specific sequences.
- Length and Melting Temperature (Tm): Usually 18–25 nucleotides long with Tm around 58–60°C for optimal amplification.
- Product Size: Amplicons typically range between 80–200 base pairs to ensure efficient and accurate qPCR amplification.
- Avoid Secondary Structures: Primer sequences should minimize hairpins or dimers to improve reaction efficiency.
Applications of GFP qPCR Primers
GFP qPCR primers are widely used in:
- Quantifying transgene expression in genetically modified organisms or cell lines.
- Monitoring promoter activity linked to GFP reporter constructs.
- Assessing gene silencing or knockdown efficiency when GFP tags are involved.
Examples and Resources
Common GFP qPCR primer sequences include:
| Primer | Sequence (5′ – 3′) |
|---|---|
| Forward | GACGTAAACGGCCACAAGTTC |
| Reverse | AAGTCGTGCTGCTTCATGTG |
Many commercial providers supply validated GFP primers for qPCR compatible with SYBR Green or probe-based assays to streamline experimental setup.
Optimizing qPCR with GFP Primers
Validation steps include:
- Testing primer efficiency with a standard curve spanning template dilutions.
- Confirming the specificity through melt curve analysis or gel electrophoresis.
- Ensuring no amplification in negative controls lacking GFP template.
Proper primer design and validation reduce errors and improve the accuracy of gene expression data using GFP as a reporter.
Key Takeaways
- GFP qPCR primers target the GFP gene to quantify expression in molecular studies.
- Optimal primers are specific with balanced melting temperatures and short amplicon sizes.
- qPCR assays with GFP primers monitor transgene expression and promoter activity.
- Primer validation ensures reliable, reproducible results in gene expression analysis.
What are GFP qPCR primers used for?
GFP qPCR primers amplify the green fluorescent protein gene in quantitative PCR. They help measure gene expression levels and track GFP-tagged proteins in samples.
How can I design effective GFP qPCR primers?
Choose primers with 18-25 bases, 50-60% GC content, and avoid secondary structures. Target a unique GFP region and keep amplicons 70-150 bp for efficient qPCR.
Which factors impact GFP qPCR primer specificity?
- Primer length and melting temperature
- Sequence uniqueness to GFP
- Avoidance of primer-dimers and hairpins
- Proper annealing temperature settings
Can GFP qPCR primers be used across different GFP variants?
It depends on sequence homology. Primers designed for standard GFP may not match variants well. Verify primer binding sites before use.
Why is amplicon size important in GFP qPCR?
Smaller amplicons (70-150 bp) amplify more efficiently in qPCR. Larger fragments can reduce reaction sensitivity and accuracy.




Leave a Comment