Is It Possible to Go from Nucleotides to Amino Acids Using BLAST?
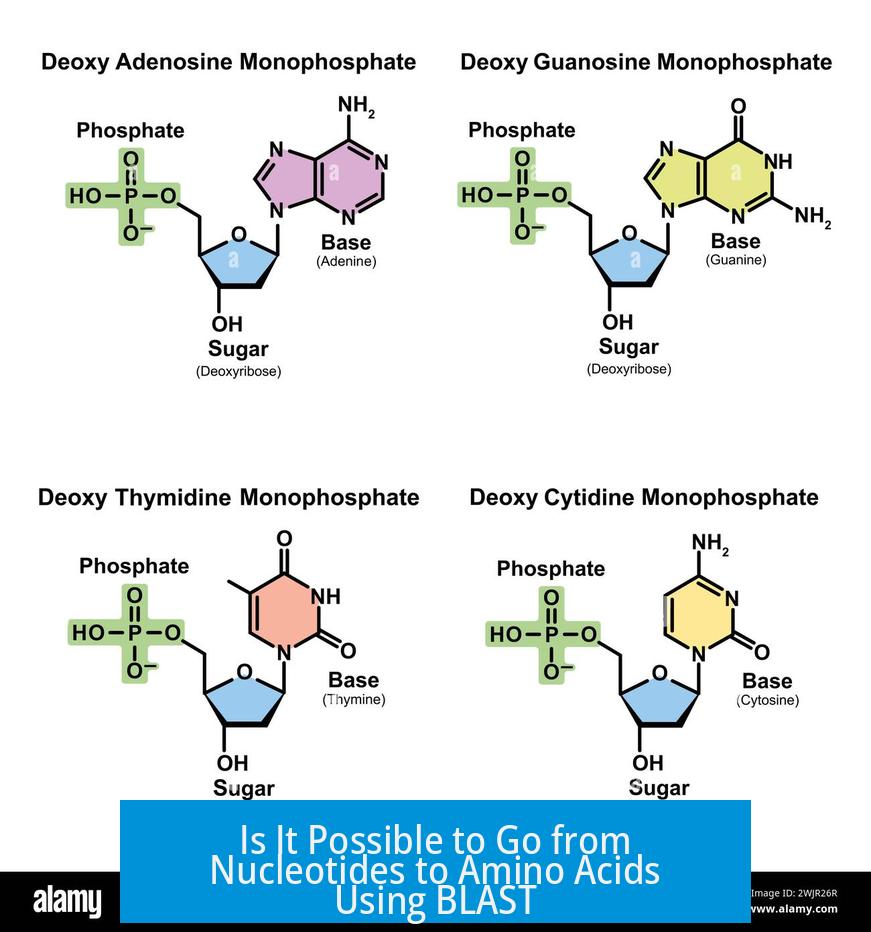
Yes, it is possible to translate nucleotide sequences into amino acid sequences using BLAST through the BLASTX program. BLASTX translates a nucleotide query in all six reading frames and compares the resulting protein sequences against a protein database. This tool helps identify potential protein products encoded by nucleotide sequences.
Using BLASTX for Nucleotide to Amino Acid Translation
The BLASTX tool is specifically designed for this purpose. Users input a nucleotide sequence, which BLASTX translates in all frames without requiring prior knowledge of the correct reading frame. Then, it aligns these translated sequences against known protein databases, returning matches with annotations. This process facilitates identification of putative amino acid sequences corresponding to the nucleotide query.
Access to BLASTX is available via the NCBI BLAST web server at NCBI BLASTX. The tool also provides alignment scores, frames used, and matching protein information.
Contextual Considerations When Using BLAST for Translation
- BLASTX results depend on the biological context, such as open reading frames (ORFs) and sequence orientation.
- It helps to have a plasmid map if analyzing sequences from plasmids, to verify the correct frame and translation start.
- Understanding protein tags and positioning (e.g., C-terminal GFP tags) improves interpretation of BLAST alignments.
Alternative Tools for Direct Sequence Translation
For straightforward nucleotide-to-amino acid translation without searching databases, dedicated tools like ExPASy Translate offer direct conversion in all frames. Users can then run sequence alignments separately to confirm orientation or homology.
When to Use BLAST for This Purpose
While BLASTX is powerful in identifying proteins related to nucleotide queries, directly blasting to translate sequences may be unnecessary if the exact coding sequence and reading frames are already known. BLAST is most useful for exploratory sequence annotation and frame determination rather than routine translation.
Key Takeaways
- BLASTX translates nucleotide sequences into amino acid sequences and aligns them to protein databases.
- Proper interpretation requires consideration of ORFs, sequence orientation, and biological context.
- Direct translation tools like ExPASy Translate exist for simple frame translation without database search.
- BLAST is better suited for identifying unknown protein-coding regions or validating annotations than routine sequence translation.
- Use plasmid maps and knowledge of sequence features to enhance accuracy when analyzing BLAST results.



Leave a Comment