Primer Specificity Checker: Tools and Best Practices
 A primer specificity checker is essential for verifying that PCR primers bind only to the intended target sequences, avoiding off-target amplification. These checks improve experiment reliability by minimizing non-specific amplification.
A primer specificity checker is essential for verifying that PCR primers bind only to the intended target sequences, avoiding off-target amplification. These checks improve experiment reliability by minimizing non-specific amplification.
NCBI Primer-BLAST Tool
The NCBI Primer-BLAST is a widely recommended tool for primer specificity checking. It allows users to input primer sequences and search for potential off-target binding sites in genomic databases.
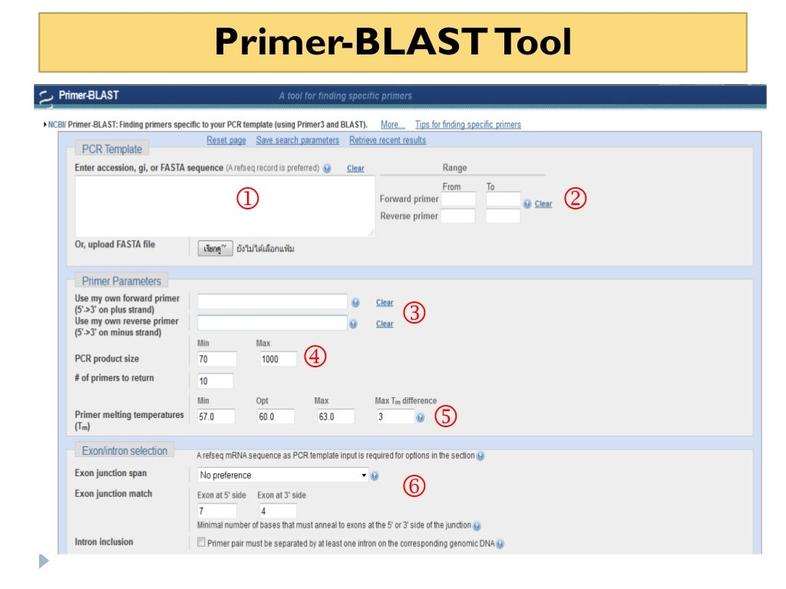
- Detects unintended binding sites.
- Supports various organisms from NCBI databases.
- User-friendly web interface: Primer-BLAST
This tool is highly reliable and often suggested by researchers for initial specificity screening.
ApE (A Plasmid Editor) for Visual Primer Analysis
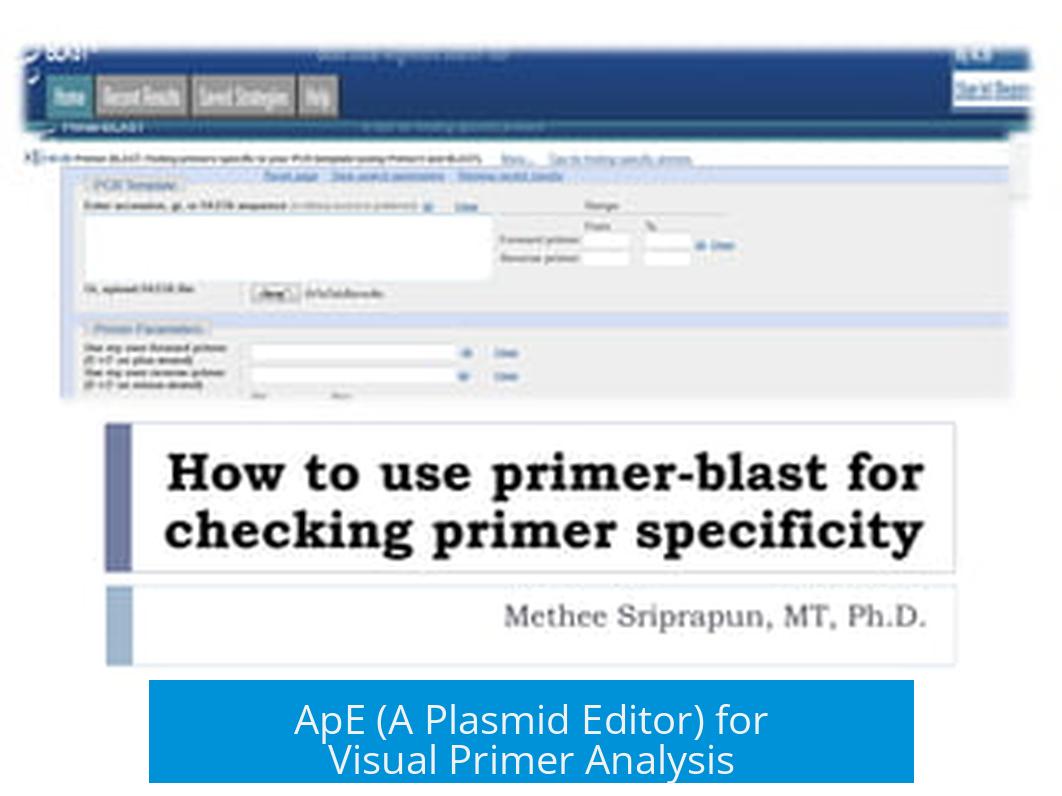
ApE is useful for analyzing primer binding on plasmid sequences. It visualizes all possible sites where primers can bind, including imperfect matches when mismatches are allowed.
- Visualizes primer locations on plasmid maps.
- Allows adjustable mismatch tolerance to find similar sequences causing off-target amplification.
- Download: ApE Software
Alternative Tools
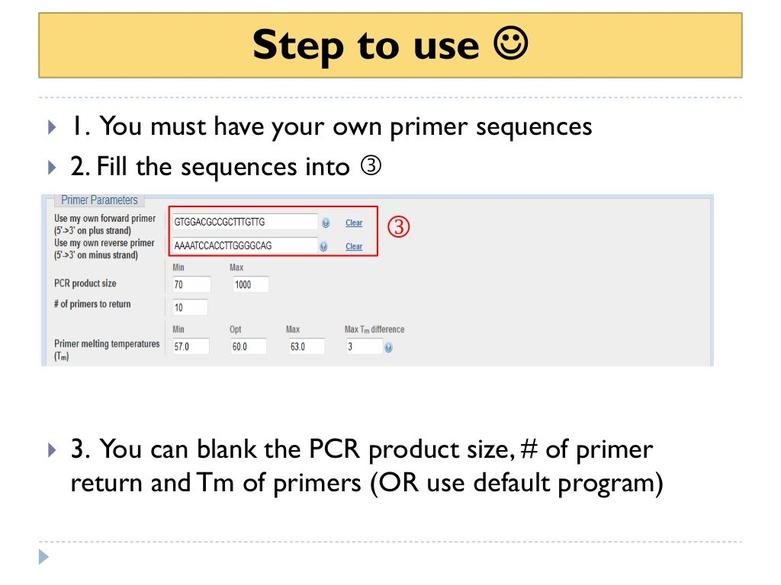
Other useful tools include:
- Benchling: Provides easy and visual sequence alignment checks for primers.
- Serial Cloner: Automates searching forward and reverse complement sequences, aiding strand-specific primer binding analysis.
- Microsoft Word: Basic text search can identify primer sequences but lacks strand orientation automation.
Practical Recommendations
Relying solely on computational specificity checks may not be sufficient. Consider sequencing PCR products with the same primers for precise verification. This step ensures that the actual amplicon matches expectations.
For PCR cleanup, using a PCR clean-up column is often enough to purify amplicons for sequencing without gel purification, provided the PCR produces a single clean band.
When designing primers with restriction sites, add extra nucleotides 5’ to the sites. This facilitates effective enzyme digestion, as some enzymes require flanking bases outside their recognition sites.
Key Takeaways
- NCBI Primer-BLAST is a primary tool to check primer specificity across genomes.
- ApE visualizes primer binding on plasmids, including mismatch tolerance.
- Benchling and Serial Cloner offer user-friendly alternatives for alignment and binding site analysis.
- Sequencing PCR products provides definitive validation beyond in silico checks.
- Use PCR clean-up columns instead of gel purification if the band is clean.
- Add extra bases at 5’ ends of primers to improve restriction enzyme digestion efficiency.
Primer Specificity Checker? A Detailed Guide to Ensuring Your Primers Hit the Right Mark
In the world of molecular biology, designing primers is like sending a letter—you want it to reach the right address without getting lost or delivered to someone else. So, what exactly is a primer specificity checker? It’s a tool that helps verify whether your primers will bind only to your intended DNA target sequence, avoiding off-target binding that could muddle your results.
Primer specificity is a dealbreaker in PCR (Polymerase Chain Reaction) and other DNA amplification techniques. If primers bind to unintended regions, you end up amplifying the wrong DNA fragments, leading to false positives or confusing data. This post dives into popular tools and clever methods to check primer specificity and improve your experiment’s accuracy.
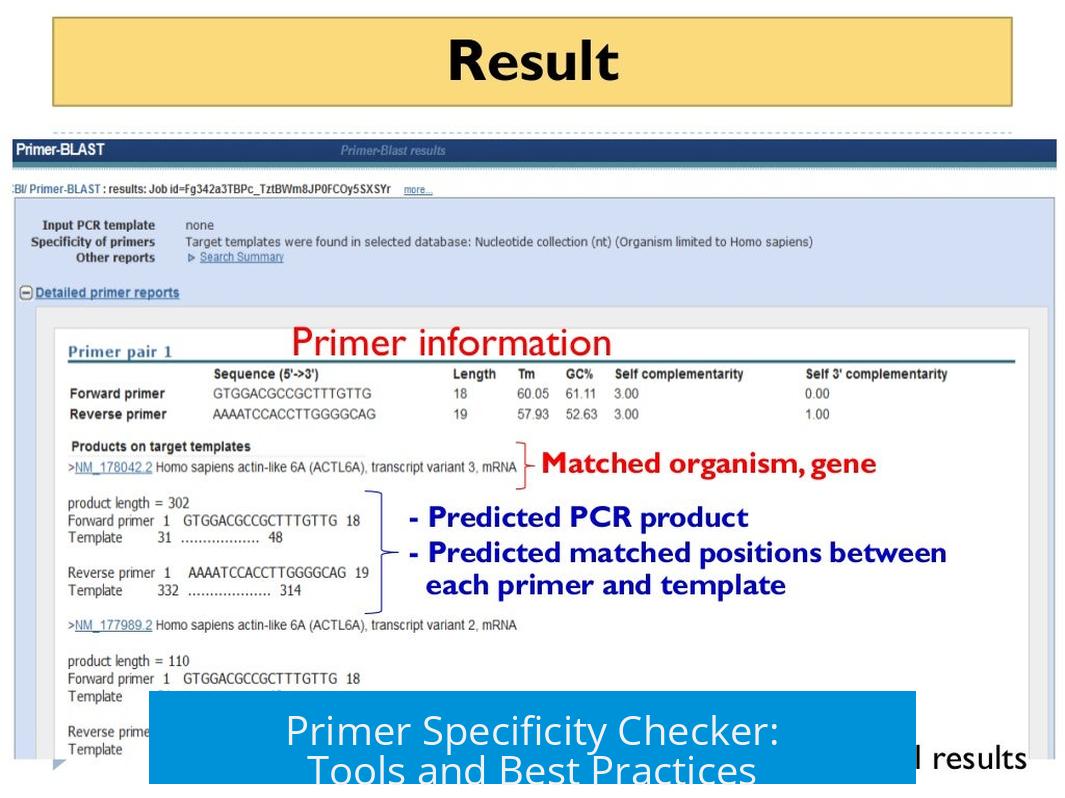
NCBI Primer-BLAST: The Gold Standard for Checking Off-Target Binding Sites
Scientists widely praise the NCBI Primer-BLAST tool for its ability to check primers against entire genomes. Here’s the scoop: this tool compares your primer sequences against a selected database, uncovering any potential off-target binding sites that might ruin your experiment.
If you’re guessing it’s a must-use tool—you’re spot on! Researchers recommend it because it not only highlights where the primers perfectly bind but also spots partial matches. This feature is important since partial primer binding can sometimes produce unexpected PCR products.
The web interface is user-friendly. You paste your primer sequences, select the organism or genome, and hit ‘blast.’ Within moments, it spits out a list showing where your primers might bind, allowing you to tweak sequences or redesign them before ordering.
Visualize Primer Binding Like a Pro With ApE (A Plasmid Editor)
Once you’ve designed primers, visual confirmation is a smart move. Imagine seeing all possible spots where your primer could stick on a plasmid. ApE (A Plasmid Editor) lets you do just that!
This handy software lets you input your plasmid’s full sequence. Then you add your primers to see exact binding locations. You can even increase the allowed mismatches to spot less-than-perfect matches that might cause unwanted amplification.
Visualizing your primer binding sites helps anticipate unexpected results, like if your primer binds in multiple locations, creating multiple bands in PCR gels. ApE makes it easier to troubleshoot before you run that expensive experiment.
Other Tools: Benchling, Serial Cloner, and Surprisingly, Microsoft Word?
Don’t limit yourself! While you may have heard about NCBI Primer-BLAST and ApE, there are other practical tools.
- Benchling: This online platform offers a slick, user-friendly interface for DNA sequence alignment and primer checking. Its visual alignment tools help you quickly scan for primer binding without headaches.
- Serial Cloner: It’s a free desktop program that automatically searches for primer matches, including reverse complements. Meaning, it knows your primer could bind reading 5’ to 3’ or the opposite—always smart to check.
- Microsoft Word? You read that right. Surprisingly, some labs copy the plasmid sequence into Word and use the search function to hunt for primer matches. While it’s not precise or scientific rigor, it’s a quick-and-dirty hack if you’re stuck without specialized software.
Sequencing Your PCR Product: The Ultimate Specificity Confirmation
Even with all these tools, nothing beats sequencing your amplified DNA for confirming your primer’s specificity. Someone suggested, “Why not just sequence the amplicon with the same primers?” It takes a few days, but it’s the direct way to know exactly what you amplified.
Many assume they need gel purification or cloning before sequencing, but that’s not always the case. If you get a clean single band from PCR, a simple PCR cleanup column removes leftover primers, nucleotides, and enzymes—making your product sequencing-ready.
This approach saves time, money, and confusion in the long run. Who wouldn’t want exact proof their primers worked perfectly?
Practical Tips: Digestion and Gel Purification Linked to Primer Design
Here’s a nugget often overlooked: if you design primers with restriction enzyme sites at their ends for cloning, remember some enzymes won’t cut effectively if the recognition site is right at the DNA fragment’s end.
Tip: Add extra nucleotides (a few bases) before the restriction site in your primer’s 5’ region. This buffer boosts enzyme cutting efficiency. Also, it’s better to digest your PCR product first, then purify it using gel extraction or a cleanup column. It’s more efficient and preserves your DNA.
Why All This Matters?
Primer specificity checker tools and methods save you from chasing shadows in your experiments. Off-target primers cause wasted reagents, inconclusive data, and countless hours of frustration.
With tools like NCBI Primer-BLAST, ApE’s visualization, and Benchling’s alignment, plus practical tips on sequencing and enzyme digestion, you gain control over your experimental fate. You’ll design primers that consistently amplify the right targets, boosting your lab’s productivity and confidence.
So, Ready to Unlock Perfect Primer Design?
What primer specificity tool suits your lab style? Do you prefer web-based solutions like NCBI Primer-BLAST, or are you more visual with ApE and Benchling? Have you ever tried the Microsoft Word trick and lived to tell the tale? Share your experiences or questions below.
Remember, the best science combines smart tools with sharp thinking. Happy primer hunting!
What is the best tool to check primer off-target binding sites?
NCBI Primer-BLAST is widely recommended for this purpose. It identifies potential off-target sites where primers may bind unexpectedly. This helps ensure primer specificity before experiments.
How can I visualize primer binding sites on a plasmid?
Use ApE (A Plasmid Editor) to load your plasmid sequence. It shows exact primer binding locations along with mismatches. This visualization aids in spotting unintended binding sites.
Are there any alternatives to NCBI Primer-BLAST for checking primer specificity?
Benchling is a good option for easy and visual alignment checks. Serial Cloner also helps, especially since it auto-searches for reverse complements of primers on sequences.
Should I rely solely on primer specificity tools for validation?
Sequencing PCR products yields exact results. Specificity checkers guide design but sequencing confirms the actual amplicon identity. It takes a few days but improves accuracy.
What is the recommended process after PCR regarding primer cleanup?
Skip gel purification if you have a single clear band. Use a PCR cleanup column to remove leftover primers and nucleotides. This streamlines preparation for sequencing or downstream applications.



Leave a Comment