Understanding Reading Frames and Open Reading Frames (ORFs)

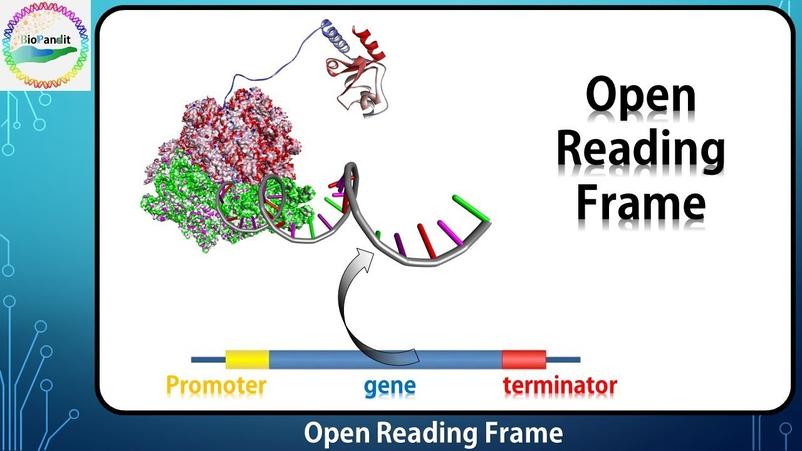
An open reading frame (ORF) is a segment of DNA or RNA that begins with a start codon and extends without interruption by a stop codon for a length sufficient to encode a protein. The key difference between an ORF and a reading frame lies in the presence of start and stop codons within the reading frame that make the ORF a realistic candidate for protein coding.
What is a Reading Frame?
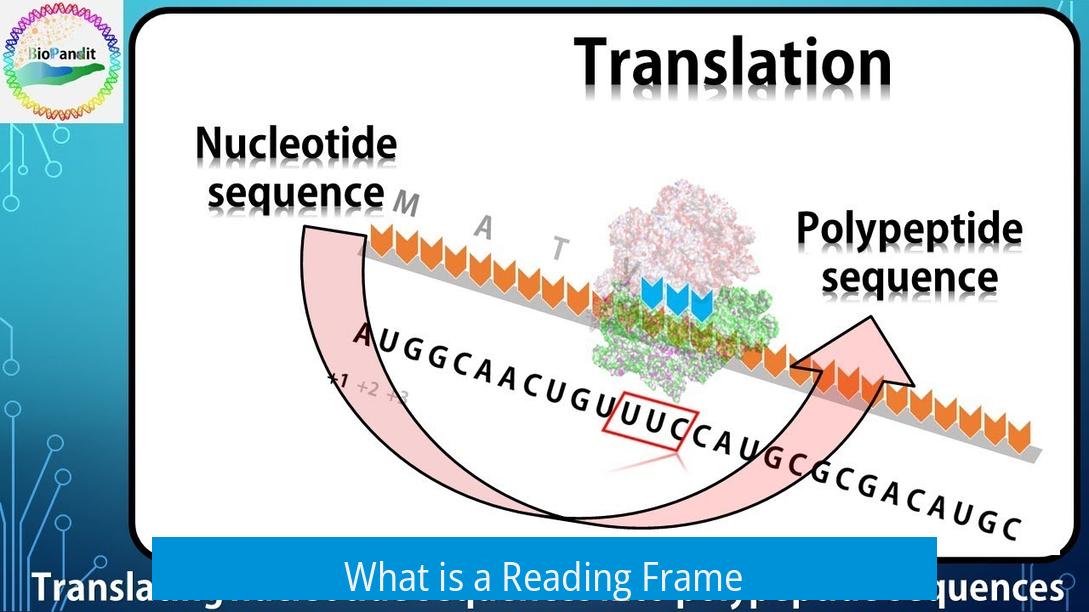
A reading frame is the way nucleotides in a DNA or RNA sequence are grouped into consecutive triplets called codons. Each codon corresponds to an amino acid or a signal during protein synthesis. Because nucleotide sequences are linear, there are three possible ways to read triplets, depending on the starting nucleotide.
- Frame 1 starts at nucleotide 1
- Frame 2 starts at nucleotide 2
- Frame 3 starts at nucleotide 3
This means any sequence has three possible reading frames. Each frame defines a different set of codons and can encode different peptide sequences.

What Defines an Open Reading Frame?
An open reading frame is a reading frame that contains a “start codon” (commonly ATG in DNA or AUG in mRNA) near its beginning and extends without encountering a “stop codon” (such as TAA, TAG, or TGA in DNA) for a length long enough to potentially encode a protein. The ORF ends at the first in-frame stop codon after the start.
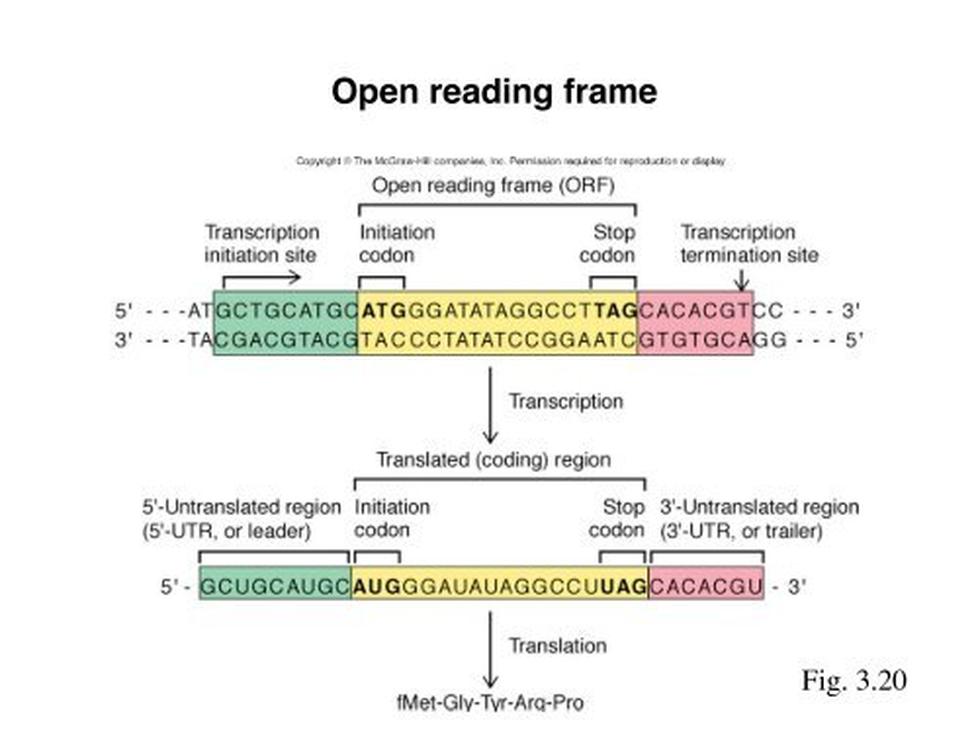
Unlike a generic reading frame, which simply partitions nucleotide sequences, an ORF is functionally significant as a potential protein-coding region. It marks the stretch of nucleotides the ribosome would translate into an amino acid chain.
Key Differences Between a Reading Frame and an Open Reading Frame
| Aspect | Reading Frame | Open Reading Frame (ORF) |
|---|---|---|
| Definition | One of three possible ways to divide a nucleotide sequence into codons. | A reading frame with a start codon and a continuous sequence without stop codon until a stop codon is reached. |
| Presence of Start Codon | Not required. | Required near the beginning. |
| Presence of Stop Codon | May or may not be present anywhere in frame. | Absent until the terminal region, marking the end of the ORF. |
| Biological Significance | Defines how the ribosome reads codons. | Predicts actual protein-coding regions. |
Context in Gene Expression
The ribosome reads the mRNA transcript codon by codon during translation. The RNA polymerase transcribes DNA into mRNA. Detecting ORFs within DNA sequences is an essential step in gene annotation since ORFs correspond to parts of the genome that could encode proteins.
Mutations, insertions, or deletions can shift reading frames, changing downstream codon groupings and potentially disrupting ORFs. Identifying the correct ORF among the three frames requires examining all frames and locating those with appropriate start and stop codons to infer coding sequences.
Summary of Key Points
- A reading frame is any one of three nucleotide codon groupings in a DNA or RNA sequence.
- An open reading frame is a reading frame beginning with a start codon and containing a continuous stretch without internal stop codons, ending with a stop codon.
- The ORF represents the likely protein-coding region readable by the ribosome.
- All sequences have three reading frames, but not all contain valid ORFs.
- Gene prediction relies on identifying ORFs within nucleotide sequences to define possible proteins.
Can Someone Explain to Me What an Open Reading Frame Is? And What’s the Difference Between an ORF and a Reading Frame?
Let’s get straight to the point: an open reading frame (ORF) is a specific type of reading frame that starts with a start codon and continues without any stop codon for a good chunk of its length, eventually ending at a stop codon, allowing it to be translated into a protein.
Confused? Don’t worry. We’ll break it down piece by piece. Why should you care? Because understanding these concepts is crucial for fields like genetics, biotechnology, and molecular biology.
What Is a Reading Frame?
Imagine a long string of letters representing nucleotide bases: A, T, G, C. These letters are grouped in threes, called codons. Each codon corresponds to an amino acid, the building block of proteins.
A reading frame is simply the way you chunk these nucleotide sequences into sets of three, but it depends on where you start. For example, given a DNA sequence that reads 123 AAABBBCCC…, you could read:
- Frame 1: Starting at base 1 — AAA BBB CCC…
- Frame 2: Starting at base 2 — AAB BBC CC…
- Frame 3: Starting at base 3 — ABB BCC C…
Each frame gives a different set of codons, and thus a totally different potential protein. So, every sequence has three possible reading frames on each strand of DNA.
The ribosome is the molecular machine that “reads” these triplet codons during protein synthesis. How it reads—reading frame—determines which amino acids are added and in what order.
But What Exactly Is an Open Reading Frame (ORF)?
Not every reading frame is useful. Some are jumbled, starting or stopping randomly. Here’s where an open reading frame enters the scene.
An ORF is a continuous stretch within a reading frame that starts with a start codon, usually ATG (or AUG in mRNA), and goes on without hitting any stop codon until it eventually encounters one. This signals the ribosome to stop translating.
Visualize it like a well-marked highway:
- Start codon = the on-ramp onto the protein-building highway.
- No stop codons along the way = no construction zones or closed lanes blocking progress.
- Stop codon = your exit, signaling the end of the protein.
For example, take the DNA snippet: ATG TTT CCA AAC…. This is an ORF if it starts near the beginning without any stop codons until the end. Because the ribosome begins at the ATG codon, it translates the sequence into a chain of amino acids uninterrupted by a stop signal.
So, What’s the Difference Between an ORF and a Reading Frame?
Here’s the lowdown:
- Reading frame is any one of the three ways you divide the nucleotide sequence into codons. It doesn’t imply anything about start or stop codons; it’s just a way of reading the sequence.
- Open Reading Frame (ORF) is a special reading frame that contains a start codon near the beginning and no stop codons for a decent distance, meaning it’s likely to be translated into a protein.
In other words, all ORFs are reading frames, but not all reading frames are ORFs. If you think of reading frames as possibilities, the ORF is the one with the green light to be a gene and make a protein.
It’s not always crystal clear which ORF corresponds to a functional protein. Sometimes you have to “translate” all three reading frames to see which one meets the criteria: starts with ATG, continues without stop codons, and ends properly.
Why Does This Matter?
If you’re a researcher, bioinformatician, or student, this knowledge is gold. It helps in gene prediction — identifying where genes are in DNA sequences. Tools that scan genomes look for ORFs to pinpoint potential protein-coding regions.
Here’s a nifty tip: keep in mind that mutations can shift reading frames (frameshift mutations) by adding or deleting nucleotides. This messes up normal reading frames and ORFs, altering protein products. A single “C” added can totally scramble the sequence, changing the meaning entirely.
And remember: it’s the polymerase enzyme that transcribes DNA into mRNA copying our sequence, and it’s the ribosome that reads those codons to make proteins. Each process depends on reading frames and ORFs to function correctly.
Real-Life Example: How ORF Discovery Drives Innovation
In biotechnology, identifying ORFs can unlock secrets to new proteins helpful in medicine, agriculture, or industry. Think about enzymes that break down plastic or proteins that fight diseases — finding the right ORFs is a key first step.
Imagine you’re decoding a mysterious DNA sequence. By scanning the three reading frames, you spot two with lots of stop codons. The third starts with ATG and runs smoothly without stops for hundreds of codons — bingo! You’ve likely found a new gene that codes for a protein.
Wrapping It Up
To recap:
- A reading frame is one of three ways to divide a DNA or RNA sequence into triplet codons.
- An open reading frame (ORF) is a reading frame that begins with a start codon and continues without stop codons for a good length, culminating in a stop codon — this is the protein-coding stretch.
- Finding ORFs helps scientists identify genes and predict proteins.
Next time you wonder what exactly an ORF is, remember: it’s the rockstar reading frame, ready to be translated into a protein. And that’s the difference between a casual reader and a protein producer.
“Genetics is like a book written in a four-letter alphabet. ORFs are the meaningful sentences waiting to be read aloud!”
Got questions? Curious about frameshift mutations or the role of start/stop codons? Drop a comment below. Learning DNA’s language is a journey — and every frame counts!




Leave a Comment