How to Confirm Gene Sub-Cellular Localization in Membranes
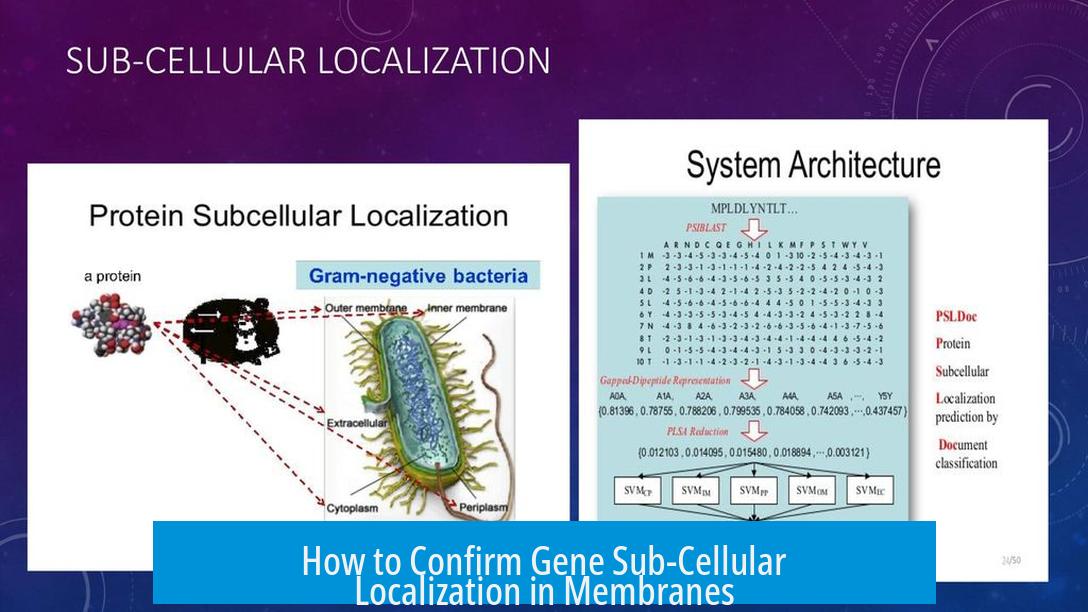
Confirming the sub-cellular localization of a gene product within the membrane involves expressing a fluorescently tagged version of the protein and visualizing its distribution using microscopy. This direct approach focuses on where the protein, not the gene’s DNA, resides in the cell.
Expressing Fluorophore-Tagged Proteins
First, researchers create a fusion protein combining the target protein with a fluorescent tag, such as green fluorescent protein (GFP). This fusion enables real-time visualization under fluorescence microscopy. The fluorescent tag reveals whether the protein localizes specifically to the cell membrane.
CRISPR/Cas-Based Targeted Labeling
Another precise method uses CRISPR/Cas technology to insert a fluorescent marker gene into the endogenous locus of the target gene. This strategy expresses the protein with its native regulatory elements intact, minimizing artifacts from overexpression.
The system used depends strongly on the organism or cell type (e.g., cultured cells, plants, mice). CRISPR editing allows in situ tagging for physiological relevance.
Adapting to Biological Systems
The choice of approach varies by experimental model. For example, in plant cells, plasmid transfection mediated by polyethylene glycol (PEG) is common to express GFP-tagged proteins. In animals or cultured cells, viral vectors or direct genome editing may be preferred.
Fluorescent fusion proteins must retain proper folding and membrane targeting to provide valid localization data.
Clarifying Protein Versus Gene Localization
It is important to focus on the protein’s localization rather than the gene DNA. The protein’s position within organelles or membranes defines the functional location relevant to cellular processes.
Additional References
- Springer Protocol Paper: Describes experimental methodology in plant systems.
- Frontiers in Plant Science Article: Provides detailed techniques for protein localization.
Summary of Key Points
- Express fluorescently tagged protein constructs to observe membrane localization.
- Use CRISPR/Cas for precise endogenous tagging of target proteins.
- Adapt methods based on the biological system (plants, animals, cultured cells).
- Focus on protein localization, not the gene DNA.
- Confirm proper folding and function of tagged proteins.
How can I visualize the membrane localization of a protein encoded by a gene?
Express a fusion protein tagged with a fluorescent marker like GFP. This allows observation of the protein on the membrane using fluorescence microscopy.
Is CRISPR/Cas useful for confirming protein localization in membranes?
Yes, CRISPR/Cas can insert a fluorescent tag at the gene’s native site. This produces a fluorescent protein for direct localization in living cells or organisms.
Does the biological system affect how localization is confirmed?
Techniques vary between systems like cells, plants, flies, and mice. Choose the method that best fits your experimental model.
When confirming gene “localization,” what exactly is localized?
Localization refers to the protein product’s position within the cell, not the gene’s DNA itself.
How is protein localization studied in plant cells?
Fluorescent-tagged proteins can be introduced via plasmid transfection using PEG. This enables membrane localization studies under fluorescence microscopy.



Leave a Comment