Understanding mRNA Decay
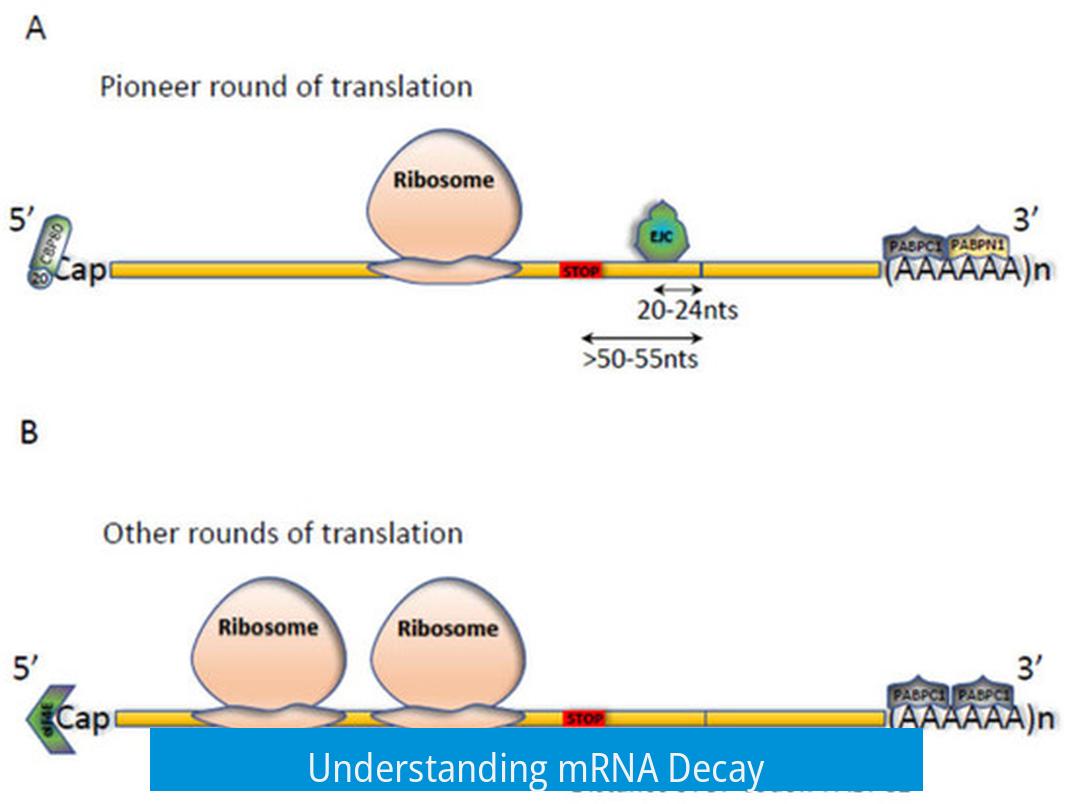
mRNA decay is a crucial cellular process that controls the stability and lifespan of messenger RNA molecules by removing specific features which initiates their degradation. This regulation ensures proper gene expression and prevents accumulation of faulty or unnecessary mRNAs.
Key Components and Initial Trigger
The process begins with the removal of the polyA tail from mRNA. This shortening destabilizes the molecule and marks it for breakdown. Without this step, mRNA remains stable and continues to be translated.
Major mRNA Decay Pathways
1. Decapping Dependent Pathway
- This pathway starts with removal of the 5’ cap structure, specifically the m7G cap.
- Enzymes DCP1 and DCP2 catalyze decapping.
- Once decapped, exonucleases XRN1 or XRN4 degrade mRNA from the 5’ to 3’ direction depending on the species.
2. Decapping Independent Pathway
- Here, the exosome complex degrades mRNA starting from the 3’ to 5’ end.
- This mechanism does not require prior decapping, distinguishing it from the first pathway.
Pathway Specificity and RNA Selection
Not all RNAs use the same decay route. Each mRNA has a preferred degradation pathway.
- Some are exclusively degraded through decapping dependent mechanisms.
- Others rely solely on the decapping independent, exosome-directed degradation.
- Some mRNAs can be degraded by either pathway.
Determining factors for this specificity are under active investigation. Research focuses on molecular signals or RNA features that influence pathway choice.
Other Influencing Factors
Additional enzymes modulate these pathways. Alternative triggers for mRNA decay include interactions with RNA interference factors like AGO proteins and siRNAs, which can guide targeted degradation.
Key Takeaways
- mRNA decay starts with polyA tail removal to destabilize the RNA.
- The two main decay pathways are decapping dependent (5’ to 3’) and decapping independent (3’ to 5’ via exosome).
- Each RNA species prefers different degradation pathways, influenced by specific signals.
- Additional proteins and RNA-based triggers can modify or induce decay.
- Understanding pathway specificity remains a significant research focus.
For more detailed information, see the referenced paper: PMC6305221.
Unlocking the Secrets of mRNA Decay: The Cell’s Recycling System
mRNA decay is the process by which cells break down messenger RNA molecules to regulate gene expression and maintain cellular health. Yes, you read that right! Just as we recycle paper to save the environment, cells recycle mRNA to keep everything running smoothly.
Understanding mRNA decay isn’t just academic babble. It means grasping how cells control which proteins get made and when. This process prevents errors, stops harmful build-ups, and even influences how plants and animals respond to stress.
Why Does mRNA Decay Matter?
Imagine mRNA as the instructions for building proteins. Once a protein is made, those instructions become outdated or might even cause harm if left unchecked. That’s where mRNA decay steps in. It signals the cell to dispose of old or faulty instructions, ensuring precision and balance.
So, what triggers this disposal? One pivotal event is the removal of the polyA tail. This long string of adenines at the mRNA’s end acts like a ‘Do Not Delete’ flag. Once snipped off, the mRNA becomes unstable, marking it ready for degradation.
The Role of the PolyA Tail: Cellular Post-It Note
Think of the polyA tail as a protective shield or a sticky note saying, “Keep me a bit longer.” When enzymes come to snip it off, it’s a cellular signal that says, “Okay, time’s up.” This removal destabilizes the mRNA molecule, making it a prime candidate for decay.
This removal is the first domino in a carefully controlled sequence of events, reassuring the cell that it won’t destroy messages prematurely. Thus, without the polyA tail, decay pathways kick in.
Which Pathway Does the Work?
The mRNA’s journey to oblivion takes two primary routes: the decapping dependent and decapping independent pathways. Picture this as two different remover teams each with their own style.
The Decapping Dependent Pathway: The 5′-3′ Express Lane
First, the mRNA’s 5′ cap, a fancy molecular hat called m7G cap, must be removed by enzymes DCP1 and DCP2. Without this cap, the mRNA is vulnerable.
Next, XRN1 or XRN4 enzymes (species dependent) swoop in, acting like a paper shredder, breaking down the mRNA from the 5’ end moving toward the 3’ end. It’s a swift and effective demolition.
The Decapping Independent Pathway: The Exosome’s Backdoor
Some mRNAs skip the decapping drama entirely. Instead, the exosome complex degrades from the opposite side, nibbling the mRNA down the 3’ to 5’ end.
This pathway operates without needing the cap removed. So, both routes are vital, but they serve different subsets of mRNAs.
Which Pathway Handles Which mRNA? The Great Specificity Puzzle
Here’s where it gets fascinating. Not all mRNAs use the same pathway. No universal recipe applies here. Some messages strictly follow the decapping dependent route; others prefer the exosome’s touch. Some are even flexible, accommodating both.
This variety sparks intense research—including work from my own lab—to uncover what decides an mRNA’s decay destiny. Is it sequence, structure, or maybe the company the mRNA keeps inside the cell? These questions keep molecular biologists buzzing.
Other Players in the mRNA Decay Ensemble
Don’t imagine this as a two-player game. Numerous other enzymes and factors influence the decay process. For example, proteins like Argonaute (AGO) and small interfering RNAs (siRNAs) can trigger mRNA degradation by targeting it specifically. This mechanism serves as a precise ‘molecular assassination’ rather than a general clean-up.
These additional triggers illustrate how versatile and regulated the decay machinery is. It’s not just random bulk destruction; it’s a targeted operation ensuring cells maintain control every step of the way.
Why Bother Understanding mRNA Decay in Detail?
Good question! Grasping mRNA decay isn’t just a biology nerd’s delight. It has practical implications for medicine and agriculture.
- In medicine, misregulation of mRNA decay contributes to diseases like cancer and neurological disorders.
- In agriculture, tweaking decay pathways could help develop stress-resistant crops.
- In biotechnology, controlling mRNA stability can optimize protein production, crucial for vaccines and therapeutics.
So, diving into the mechanisms of decay helps scientists craft better therapies and innovations.
For the Curious: Digging Deeper Into mRNA Decay
If you want a detailed tour through the molecular machinery, consider this valuable paper authored by experts in the field. It explores how plants manage mRNA decay, but many principles apply across organisms:
A Comprehensive Study on mRNA Decay Mechanisms
This resource nicely ties together the enzymes involved, pathways, and functional consequences, highlighting how evolution conserved this essential process.
Wrapping Up the Cellular Sermon
mRNA decay might sound like molecular garbage disposal, but it’s much more—a sophisticated quality control and regulation mechanism. The process starts with removing the polyA tail and continues through specialized pathways that vary depending on the mRNA.
This selective, multi-pathway system ensures cells control protein production with precision. Our understanding continues to grow, revealing not just how cells clean house but also potential avenues to combat disease and improve health.
Next time you think about gene expression, remember there’s a tight, elegant balance between creating and destroying—that’s the art of mRNA decay!
What triggers the start of mRNA decay?
mRNA decay begins with the removal of the polyA tail. This removal destabilizes the mRNA and signals that it should be degraded.
How does the decapping dependent pathway work?
In this pathway, enzymes DCP1 and DCP2 remove the m7G cap. Then, the exonuclease XRN1 or XRN4 degrades the mRNA from the 5′ end toward the 3′ end.
Does every mRNA use the same decay pathway?
No. Different mRNAs prefer different pathways. Some are only degraded by the decapping dependent pathway, some only by the decapping independent pathway, and some by both.
What is the role of the exosome in mRNA decay?
The exosome degrades mRNA from the 3′ end to the 5′ end. It does not need the mRNA to be decapped first to carry out degradation.
Are there other mechanisms that can trigger mRNA decay?
Yes. Other enzymes like AGO and siRNAs can trigger mRNA degradation through different routes besides the main pathways.



Leave a Comment