Single-cell Multi-omics: Explained
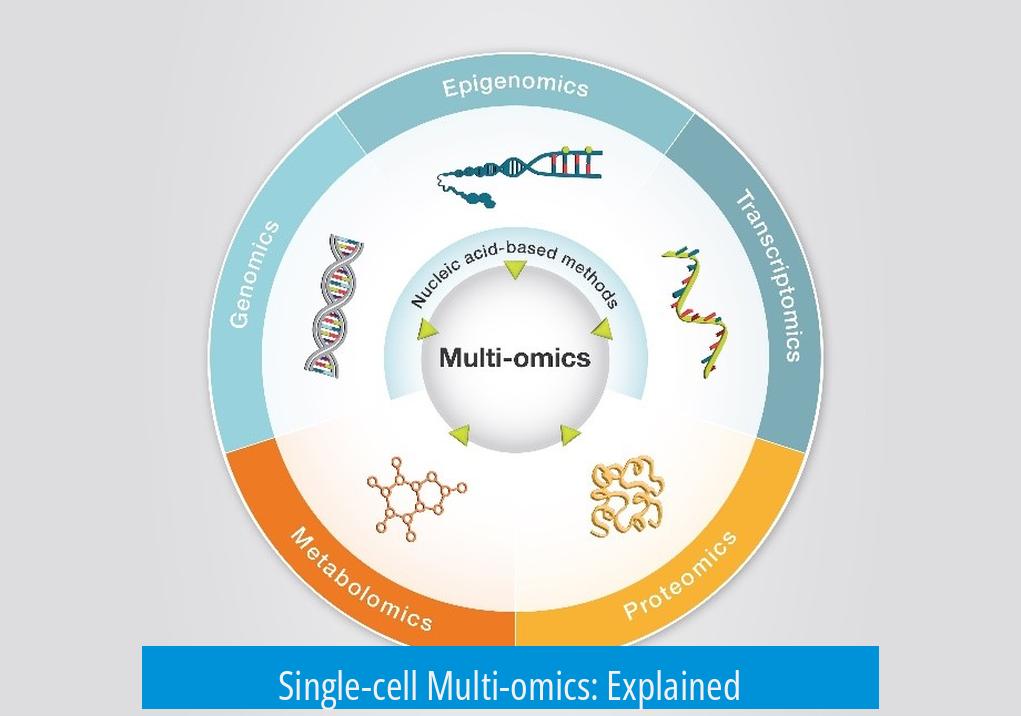
Single-cell multi-omics integrates multiple molecular analyses at the individual cell level to understand cellular functions fully, covering DNA, RNA—including noncoding RNA—and proteins. This approach overcomes limitations seen in traditional bulk sequencing by revealing cellular diversity.
From DNA to Protein: Beyond Coding RNA
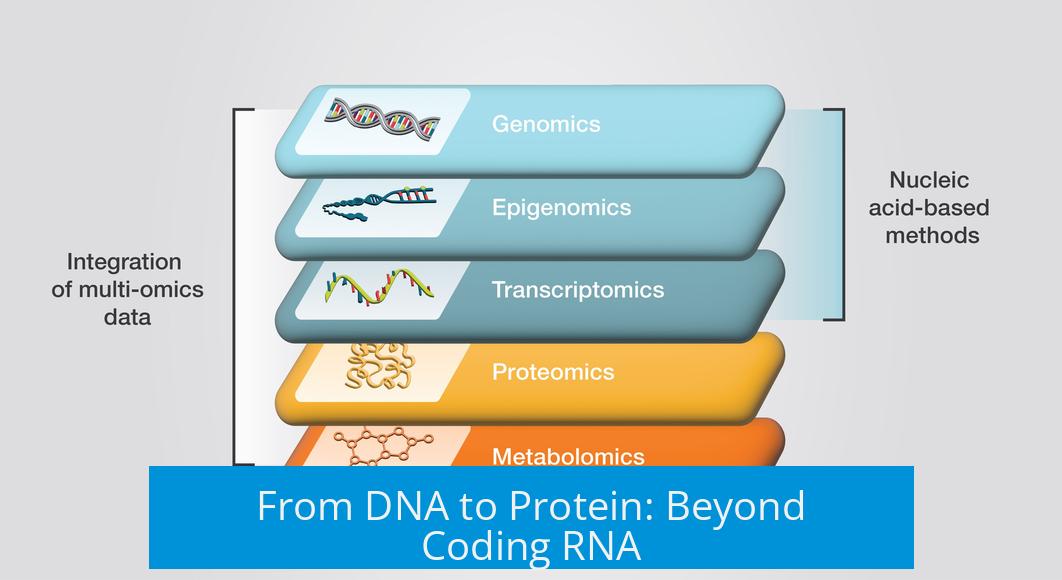
The basic biological flow describes DNA transcribed to RNA, which translates into proteins. However, this traditional framework emphasizes only coding RNA—mRNA. Single-cell multi-omics expands this view by including noncoding RNAs, integral regulatory molecules that influence gene expression without encoding proteins.
- Coding RNA (mRNA): Carries genetic instructions for protein synthesis.
- Noncoding RNA: Includes molecules like microRNA and long noncoding RNA, regulating gene expression and cell function.
This broader scope provides insight into complex regulatory networks within individual cells that coding RNA data alone cannot reveal.
Tracking Noncoding Regulatory RNA in Single Cells
Single-cell multi-omics captures the activity of these noncoding RNA species, which modulate transcription, translation, and epigenetic states. Their identification helps explain cell-to-cell variability and mechanisms driving development, disease, and response to therapy.
Researchers working with noncoding RNA highlight the method’s ability to access this previously underexplored layer of regulation, which traditional bulk techniques often obscure.
How Is There Enough Sample from a Single Cell?
Single cells contain limited material, raising questions about the feasibility of multi-omics profiling. Advances in sensitive biochemical techniques and amplification methods enable researchers to detect DNA, RNA, and proteins from minute quantities reliably.
- Amplification techniques boost nucleic acid signals.
- Proteomic methods adapted to low input samples.
- Integration across molecules from the same cell preserves context.
These technical innovations ensure robust data capture from single cells.
Appreciation of Single-cell Multi-omics
The field represents a major leap in molecular biology. It allows scientists to dissect cellular heterogeneity, understand complex tissues, and identify rare cell types or states. Such insights drive advances in development biology, cancer research, and personalized medicine.
“This is amazing stuff!”—a common sentiment among those exploring single-cell multi-omics.
Key Takeaways
- Single-cell multi-omics profiles DNA, RNA, and proteins from individual cells.
- It captures coding and noncoding RNAs, expanding understanding of gene regulation.
- Sensitive amplification techniques make analysis of single-cell samples possible.
- This technology reveals cellular diversity and mechanisms hidden in bulk data.
Single-cell multi-omics: Explained
Single-cell multi-omics is a groundbreaking approach that lets scientists explore the full complexity of individual cells by analyzing multiple molecular layers—DNA, RNA, proteins, and beyond—all at once. At first blush, you might think it’s just about reading DNA to see which genes get turned into RNA and then proteins. But the story is far richer. This technique is transforming our understanding of biology, medicine, and even how we define what a cell truly is.
Imagine trying to understand a city’s life cycle by looking only at its blueprint. That’s what looking at DNA alone is like. Sure, it tells you what’s *possible.* But single-cell multi-omics dives deep into the activities, revealing who’s working, who’s resting, and who’s planning a party tonight.
Moving Beyond the Classic DNA → RNA → Protein Chain
The classic biology lesson says: DNA turns into RNA, which then makes proteins. Sounds neat and tidy, right? But it only covers coding RNA—those bits that eventually make proteins. What gets often overlooked is the vast sea of noncoding regulatory RNA.
These aren’t just random sequences or biological noise. Noncoding RNAs act like tiny cell managers and DJs, controlling when and where genes turn on without ever becoming proteins themselves. Single-cell multi-omics lets researchers capture this regulatory info, layering crucial context over the usual genetic blueprint. This insight is game-changing for diseases where regulation goes haywire, like cancer or neurological disorders.
The Unsung Heroes: Noncoding Regulatory RNAs
Think of noncoding RNAs as the unsung heroes behind the scenes. These molecules can modulate the expression of genes, shape how cells react to stress, or even orchestrate developmental programs. Typical RNA sequencing misses much of this nuance.
A scientist working with noncoding RNA famously pointed out how valuable single-cell multi-omics is for capturing these regulators. It’s like upgrading from a black-and-white image to full-color, 3D footage. By including noncoding regulatory RNA data, researchers glimpse the cell’s control panel, not just the output.
Sample Size, Meet Single Cells: How Do They Do It?
At this point, you might be scratching your head. “Wait, how in the world is there enough sample from a single cell for you to work with?” It’s a fair question. Individual cells are tiny, and collecting enough material for DNA, RNA, and protein analysis seems almost impossible.
Here’s the scoop: scientists use advanced amplification techniques that magnify these minute molecular quantities without distorting their original makeup seriously. Think of it as turning a whisper into a shout while keeping the words intact. Alongside clever microfluidics and miniaturized labs-on-a-chip, these innovations make single-cell multi-omics not just feasible but surprisingly robust.
Why Single-cell Multi-omics Is Mind-Blowingly Amazing
It’s no wonder researchers are buzzing with excitement. Single-cell multi-omics offers a view into the cellular universe that was science fiction just a decade ago. Knowing what each cell does, how it regulates itself, and what proteins it churns out means better understanding development, disease progression, and treatment responses on a personalized level.
Curious newbies to this field often find themselves deeply engaged—and a bit overwhelmed—by the complexity and promise. That’s normal! One enthusiastic learner shared they had literally just been googling “single cell” on Wikipedia before discovering this technique.
Practical Impact and Future Horizons
Scientists now use single-cell multi-omics in fields like cancer research, immunology, neurology, and developmental biology. For instance, by dissecting tumors one cell at a time, researchers can find out which cells resist chemotherapy and why, paving the way for smarter, targeted drugs. It’s like having a microscope not only to see cells but to eavesdrop on their conversations and decisions.
Moreover, the inclusion of noncoding regulatory RNAs provides clues scientists missed before. This can explain why some therapies work well for some patients but not others, moving medicine closer to the “precision” ideal.
If you’re a student or a scientist, getting familiar with single-cell multi-omics means stepping into the future. It’s complex, sure, but understanding these layers prepares you for research that’s both deep and clinically transformative.
Want to Dive In? Here’s What to Keep in Mind
- Single-cell multi-omics is *not* just a more detailed sequencing. It integrates multiple data types from one tiny cell.
- Don’t overlook noncoding RNA—they’re crucial players in gene regulation and cellular behavior.
- Sample size is tiny but manageable with modern amplification and microfluidics techniques.
- The results inform everything from basic biology to personalized medicine.
In short, single-cell multi-omics shines a spotlight on the hidden complexity of life, cell by cell. It’s a thrilling era for biology—one where we don’t just look at cells; we listen and learn from their inner workings.
What makes single-cell multi-omics different from traditional RNA studies?
Single-cell multi-omics analyzes both coding and noncoding RNA. It captures regulatory RNA that traditional DNA → RNA → protein studies often miss. This approach gives a fuller picture of cellular functions.
How does single-cell multi-omics handle the tiny amount of material from one cell?
Advanced techniques amplify and analyze small samples. They ensure enough data comes from a single cell for meaningful multi-omics study. This overcomes the challenge of limited sample size.
Why is noncoding regulatory RNA important in single-cell analysis?
Noncoding RNA controls gene expression and cell behavior. Studying it reveals mechanisms that coding RNA alone can’t explain. Single-cell multi-omics uncovers this crucial layer.
Can single-cell multi-omics study all types of RNA at once?
Yes, it measures coding and noncoding RNA simultaneously. This holistic approach helps researchers understand gene regulation and protein production in one experiment.
What benefits does single-cell multi-omics offer for research?
It delivers precise molecular data at the individual cell level. This helps identify diverse cell types and states. The method advances understanding of complex biological systems.



Leave a Comment