Allele Specific PCR Amplification: Can It Be Wrong or May It Work?
Allele-specific PCR amplification can work, but its success heavily depends on multiple factors such as the template DNA type, product size, and the nature of the SNP. In some cases, it may fail or yield ambiguous results, especially when targeting single nucleotide polymorphisms (SNPs).
Template DNA and Product Size
The choice between cDNA or genomic DNA as the template influences amplification efficiency. cDNA usually offers less complexity and fewer introns, potentially increasing specificity. Genomic DNA may complicate amplification due to intron presence and larger product size.
Product size matters; shorter amplicons (typically under 300 base pairs) tend to amplify more reliably. Longer fragments carry more risk of non-specific binding or incomplete amplification. When designing allele-specific primers, these parameters must be optimized for clean results.
Challenges of Single SNP Targeting
Allele-specific PCR aims to distinguish alleles differing by a single nucleotide at the primer binding site. However, a single SNP often does not affect primer annealing enough to prevent non-specific amplification.
Experiences show that even multiple SNPs may not guarantee allele specificity in PCR. The risk of false positives or non-specific bands increases when the mismatch is subtle. This makes designing primers and reaction conditions critical.
Alternative Approaches: Restriction Enzyme Analysis
In some cases, detecting SNPs via allele-specific PCR may be less efficient or prone to error. An alternative is to exploit whether the SNP creates or removes a restriction enzyme site.
- Amplify a region by standard PCR.
- Digest the PCR product directly with the appropriate restriction enzyme.
- Analyze digested fragments by gel electrophoresis.
This method avoids complex primer design and reduces risk of non-specific products. It also simplifies the workflow by eliminating purification steps.
Summary: Key Points to Consider
- Allele-specific PCR can work but requires careful primer design and optimization.
- Single SNPs often produce minimal differences in priming efficiency, which may lead to incorrect amplification.
- Template type and product size influence amplification success.
- Alternative methods like restriction enzyme digestion offer a practical approach when allele-specific PCR is unreliable.
Allele Specific PCR Amplification: Wrong or May Work?
Is allele specific PCR amplification a reliable method, or is it a recipe for frustration? The straightforward answer: it can work, but it’s tricky and depends heavily on several key factors. Let’s dive into the murky waters of allele-specific PCR and figure out when to brace yourself and when to celebrate success.
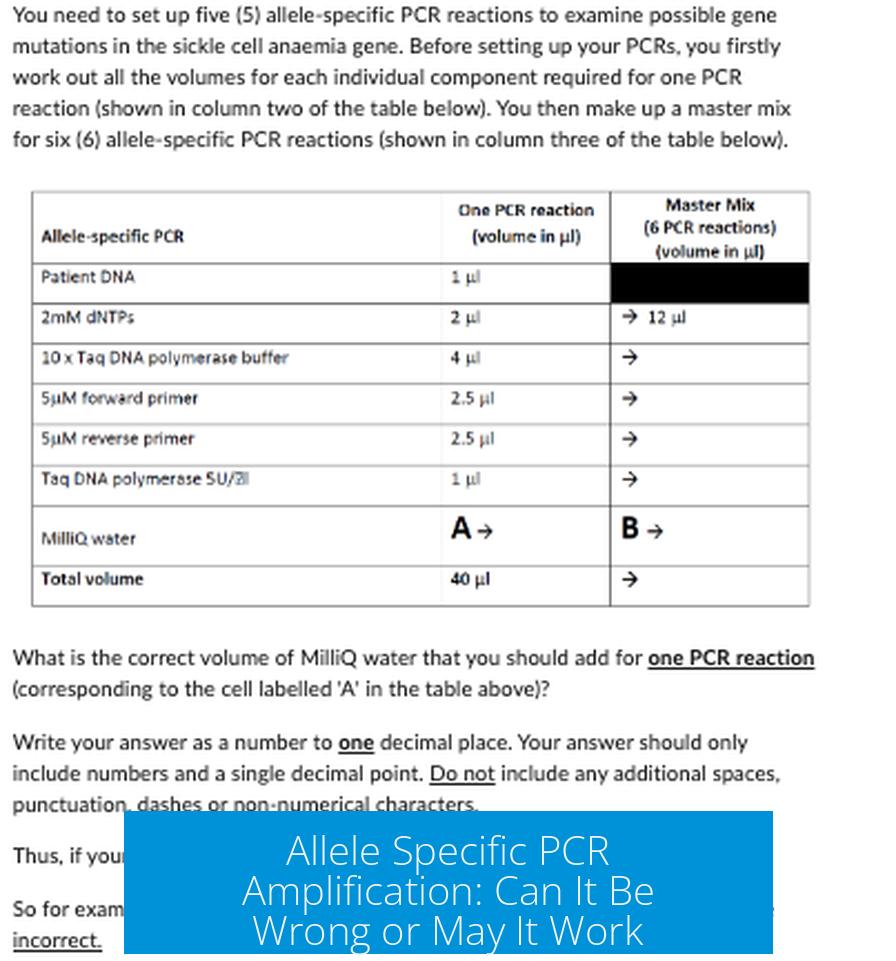
Imagine you’re trying to amplify a particular allele containing a single nucleotide polymorphism (SNP)—it sounds simple enough. But beware: that one tiny difference can either be a mere blip or a massive headache in your PCR amplification experiment.
Template DNA: The Starting Point Matters
First things first: Are you amplifying from cDNA or genomic DNA? This isn’t a trivial detail. The type of template DNA dramatically affects your PCR.
Why? Genomic DNA is usually longer, more complex, and harder to amplify due to introns and repetitive regions. In contrast, cDNA represents processed mRNA and lacks introns, often making amplification cleaner and easier.
Next is the product size. How many base pairs (bp) are you trying to amplify? Larger products tend to reduce efficiency and specificity. In allele-specific PCR, a compact target below ~300 bp is ideal to increase the odds of success.
So if your assay involves large genomic regions with heavy complexity, allele-specific PCR amplification might face an uphill battle.
Single SNPs Aren’t Always the Villains, But They Can Be Sneaky
Here’s a curious thought. One might assume that a single SNP should easily direct allele specificity in PCR. Surprisingly, it often does not.
Why? Because a single nucleotide change may not drastically alter the primer binding enough to exclude the non-target allele. PCR enzymes are quite forgiving. A single mismatch rarely kills amplification outright.
In fact, some researchers have observed that even with multiple SNPs present, allele-specific PCR can falter. Amplification differences can be subtle and inconsistent, making results ambiguous.
So, relying solely on a single SNP to distinguish alleles by PCR can sometimes be chasing ghosts.
An Elegant Alternative: SNPs and Restriction Enzymes
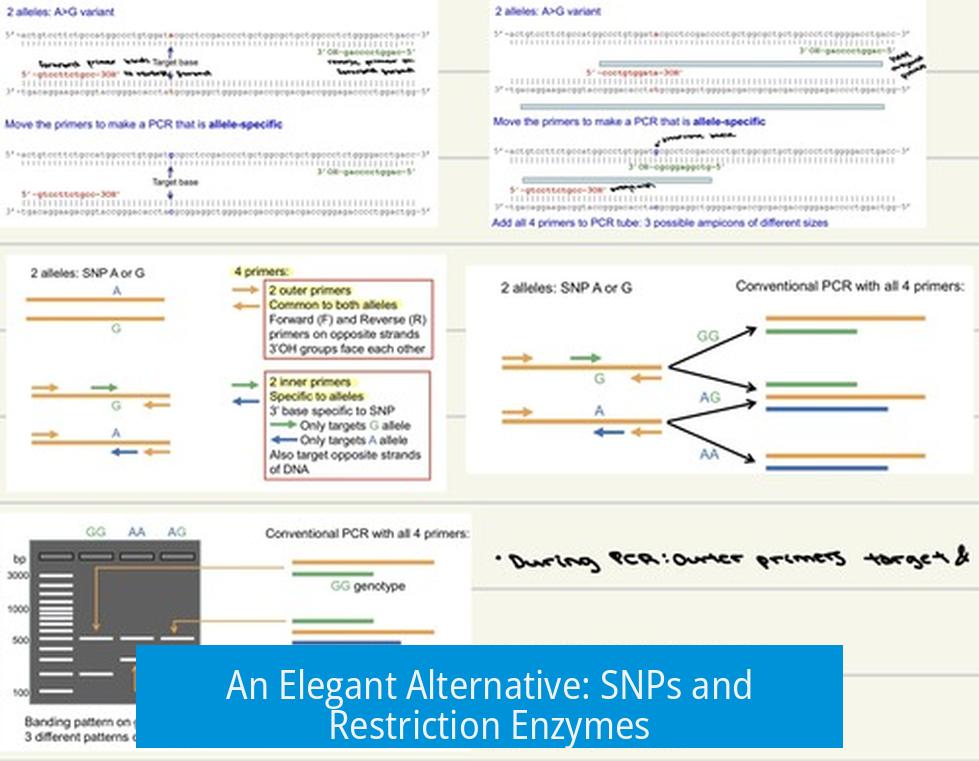
Here’s where a clever trick enters the scene. If your SNP creates or abolishes a restriction enzyme site, you can leapfrog some of the pain of allele-specific PCR.
This method involves amplifying a general region encompassing the SNP, then digesting the PCR product with the appropriate restriction enzyme.
The beauty? No need to purify the PCR product. Just mix it with the enzyme and incubate. If the SNP is present (or absent), the enzyme will cut (or fail to cut), yielding a distinct band pattern upon gel electrophoresis.
This approach tends to be more robust, cleaner, and less finicky because it doesn’t hinge on tiny differences in primer binding but instead exploits the SNP’s effect on restriction sites.
Navigating The Challenges: Tips and Tricks
- Design primers carefully. Place the SNP at the 3’ end of your allele-specific primer to increase discrimination.
- Keep the amplicon short. Aim for under 300 bp to boost efficiency.
- Validate with controls. Test your primers on known homozygous and heterozygous samples to confirm specificity.
- Consider nested PCR. A second round of amplification can enhance specificity but adds complexity.
If allele-specific PCR seems too temperamental, don’t despair. Use the restriction enzyme digestion method when SNPs influence cutting sites.
Real-World Example: When Allele-Specific PCR Worked—and When It Didn’t
In one lab, researchers tried allele-specific PCR for a SNP in a gene related to disease susceptibility. Amplification produced ambiguous bands, sometimes failing to differentiate alleles across multiple trials.
Switching to the restriction digest protocol solved their problem. The SNP happened to eliminate a EcoRI site in one allele. After PCR and enzyme digestion, the two alleles clearly displayed different band sizes on an agarose gel, making interpretation straightforward.
In another case, with cDNA as the template and small amplicons under 150 bp, allele-specific PCR worked reasonably well, probably because cDNA has fewer complexities than genomic DNA.
To PCR or Not to PCR?
Allele-specific PCR is like a precision scalpel. When used right, it cuts neatly and accurately. When misused, it’s a blunt tool that frustrates and misleads.
It’s crucial to consider:
- What template DNA you’re using.
- The size of your PCR product.
- The nature of your SNP—does it impact primer binding enough?
- Are alternative methods like restriction digestion more sensible?
Final Thoughts
Allele specific PCR amplification may work, but it’s not foolproof or universally straightforward. It demands thoughtful design, validation, and sometimes patience. The strategy can be powerful, especially with small amplicons from cDNA.
Yet, the stubborn realities of enzyme kinetics, primer binding, and genomic complexity mean one should not bank on a single SNP’s location alone to guarantee success. Keep an open mind about other approaches such as enzyme digestion, sequencing, or probe-based assays.
So the next time you ask if allele-specific PCR amplification is “wrong”—remember, it’s often not wrong, but it can be a bit like walking a tightrope. Proceed with caution, prepare backup plans, and make your plan based on detailed knowledge and practical testing.
PCR lovers and haters alike, what’s your allele-specific PCR story? Triumph or disaster? Share below!
Q1: Can allele-specific PCR fail when targeting single SNPs?
Yes, allele-specific PCR can be tricky with single SNPs. A single nucleotide change might not strongly affect amplification, making it hard to get allele-specific results in some cases.
Q2: Does the type of DNA template affect allele-specific PCR success?
Yes, whether you use cDNA or genomic DNA impacts the outcome. Product size also plays a role. Larger products or complex templates can reduce amplification efficiency.
Q3: Are there alternatives to allele-specific PCR for SNP detection?
Yes. Using restriction enzymes to detect SNPs that create or eliminate restriction sites is a simpler option. This method avoids complex allele-specific amplification steps.
Q4: Why might allele-specific PCR struggle with multiple SNPs?
Multiple SNPs can increase complexity, making primer design and selective amplification more challenging. This often lowers the specificity and reliability of the PCR.




Leave a Comment