RNA Extractions using Qiagen RNeasy Mini Kit
The Qiagen RNeasy Mini Kit enables efficient RNA extraction from various tissues, but preparatory steps and sample type critically influence the success and purity of RNA. Proper tissue processing, centrifugation conditions, and quantification method choices are essential for optimal results.
Preparation of Tissue Prior to Extraction
Before using the RNeasy Mini Kit, many researchers grind tissue directly in Trizol reagent. This step helps lyse cells thoroughly and inactivates RNases, improving RNA yield. For example, Drosophila tissues are often homogenized in Trizol before applying the RNeasy protocol.
Fatty tissues present challenges. Lipids can clog columns and reduce RNA purity. A crude purification with Trizol or a hybrid method can reduce lipid interference. The Bloomington Drosophila Stock Center provides a protocol aligning Trizol grinding with RNeasy purification, addressing such difficulties [source].
Challenges with Sample Type and Spin Columns
- Lipid-rich tissues and yolk-heavy samples, such as certain insect larvae and Xenopus embryos, often compromise column efficiency due to clogging.
- Alternatives like the Zymo Research Direct-zol kit combine Trizol extraction with a Qiagen-inspired spin column, minimizing processing time and enhancing RNA quality.
Centrifugation and Sample Handling Conditions
Room temperature centrifugation is recommended to prevent precipitate formation that can block the membrane of the spin column. Cooler temperatures might cause precipitation of salts or other compounds, hampering RNA binding and elution.
The Qiagen troubleshooting guide advises careful adherence to centrifugation parameters and buffer compositions to avoid sample loss or contamination.
RNA Quantification and Contaminants
Using NanoDrop spectrophotometers often produces unreliable RNA quantification due to contaminant interference, such as phenol or salts. Fluorescence-based assays like Qubit or Ribogreen are preferred for accurate measurements.
Monitoring 260/280 and 260/230 ratios is less critical if the extraction includes DNase treatment and follows recommended steps. Residual contaminants typically do not interfere with downstream applications after dilution.
Handling High RNA Concentrations
Highly concentrated RNA samples may exceed the measurement range of quantification instruments. Diluting samples before reading ensures accuracy and prevents saturation errors. Dilutions of 1:10 or greater often resolve this problem.
Key Takeaways
- Pre-grinding tissue in Trizol improves RNA recovery and reduces inhibitors.
- Fatty or yolk-rich tissues may require modified protocols or hybrid kits.
- Spin columns work best with room temperature centrifugation to avoid clogging.
- Fluorescent quantification (Qubit, Ribogreen) is more reliable than spectrophotometry.
- Dilute high-concentration samples to ensure accurate RNA quantification.


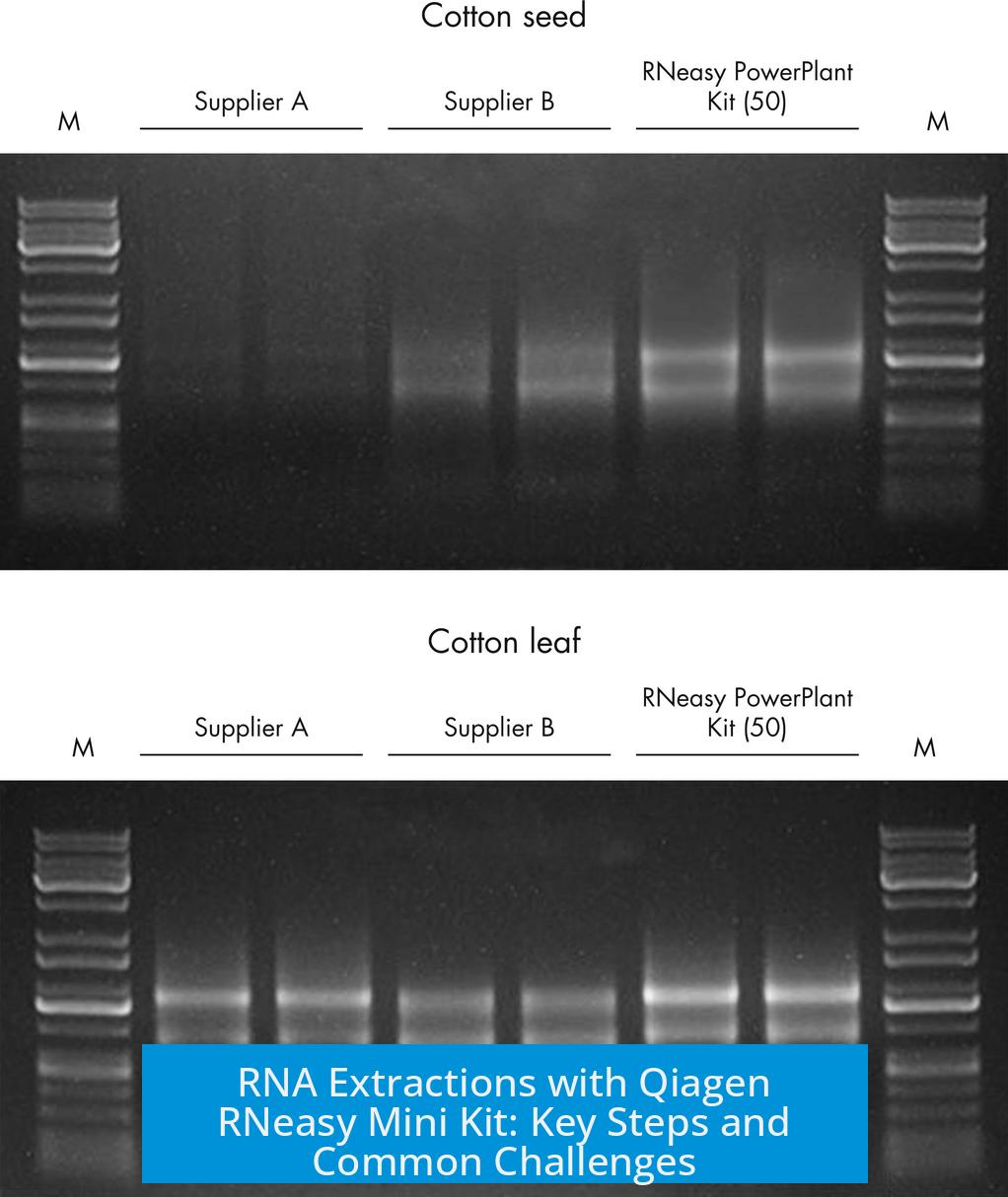

Leave a Comment