What is the “Modified Local Alignment Algorithm”?
The modified local alignment algorithm generally refers to adaptations of local sequence alignment methods, like Smith-Waterman or BLAST, designed to improve speed or sensitivity for specific tasks. It often involves adjustments to scoring schemes or heuristics for faster identification of similar regions in biological sequences.
Understanding Local Alignment Algorithms
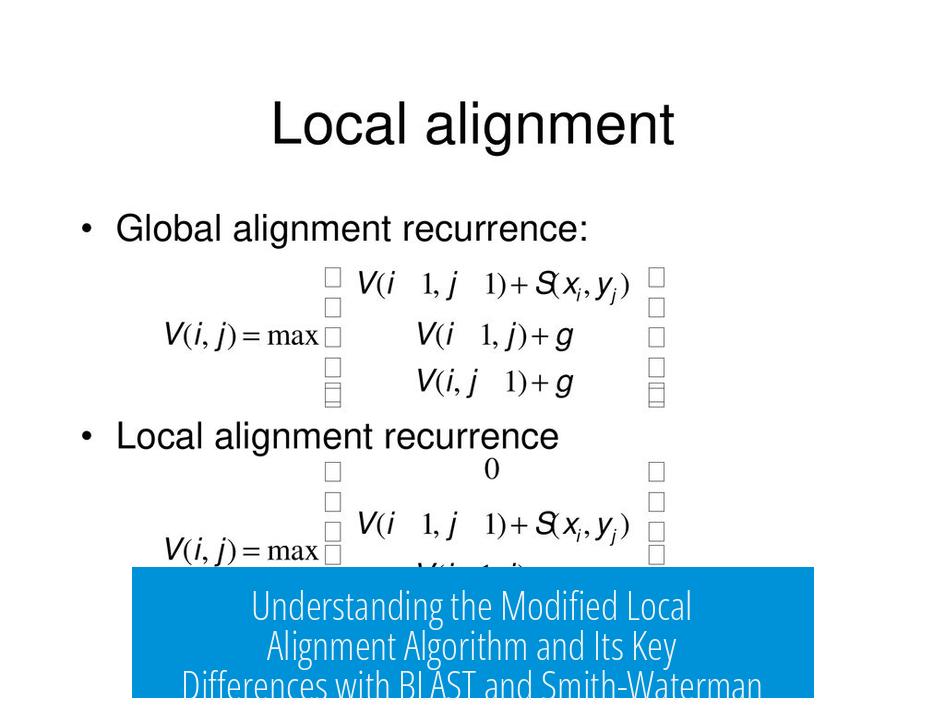
Local alignment finds the best matching region between two sequences without aligning the entire length. The Smith-Waterman algorithm is a classic example, providing optimal local alignments but at high computational cost.
BLAST (Basic Local Alignment Search Tool) represents a modified, heuristic approach. Instead of exhaustive comparison, it starts with short matching “seeds” and extends only promising alignments to improve speed.
Key Differences: BLAST vs Smith-Waterman
- Smith-Waterman: Uses dynamic programming with mismatch penalties, match scores, and affine gap penalties to find exact optimal local alignments.
- BLAST: Employs a neighborhood score threshold (T) during initial seeding to filter short sequences. This step modifies alignment by focusing on high-scoring seed matches.
This seed-based heuristic modification sacrifices exhaustive optimality for faster database searching while maintaining biologically significant alignments.
Parameters in Modified Local Alignment
Typical parameters include:
- Match score: Positive value for matching residues.
- Mismatch penalty: Negative score for mismatches.
- Gap penalties: Affine functions penalizing gap openings and extensions.
- Neighborhood score threshold (T): Unique to BLAST, this parameter controls which seeds pass to extension.
These parameters define the sensitivity and specificity of the alignment process in modified local alignment algorithms.
Applications and Purpose
Modified local alignment algorithms balance accuracy and computational efficiency in searching and comparing biological sequences. They are widely used in genomics for identifying homologous regions, annotating genes, or detecting evolutionary relationships.
BLAST, as a prime example, enables rapid screening of vast databases, which classic local alignment algorithms cannot handle feasibly.
Key Takeaways
- The “modified local alignment algorithm” often refers to heuristic adaptations like BLAST.
- BLAST uses a neighborhood score threshold to filter initial seeds for extension.
- Classic local alignments rely on match scores, mismatch penalties, and affine gap penalties.
- Modifications improve speed and efficiency while retaining biologically relevant local similarities.



Leave a Comment